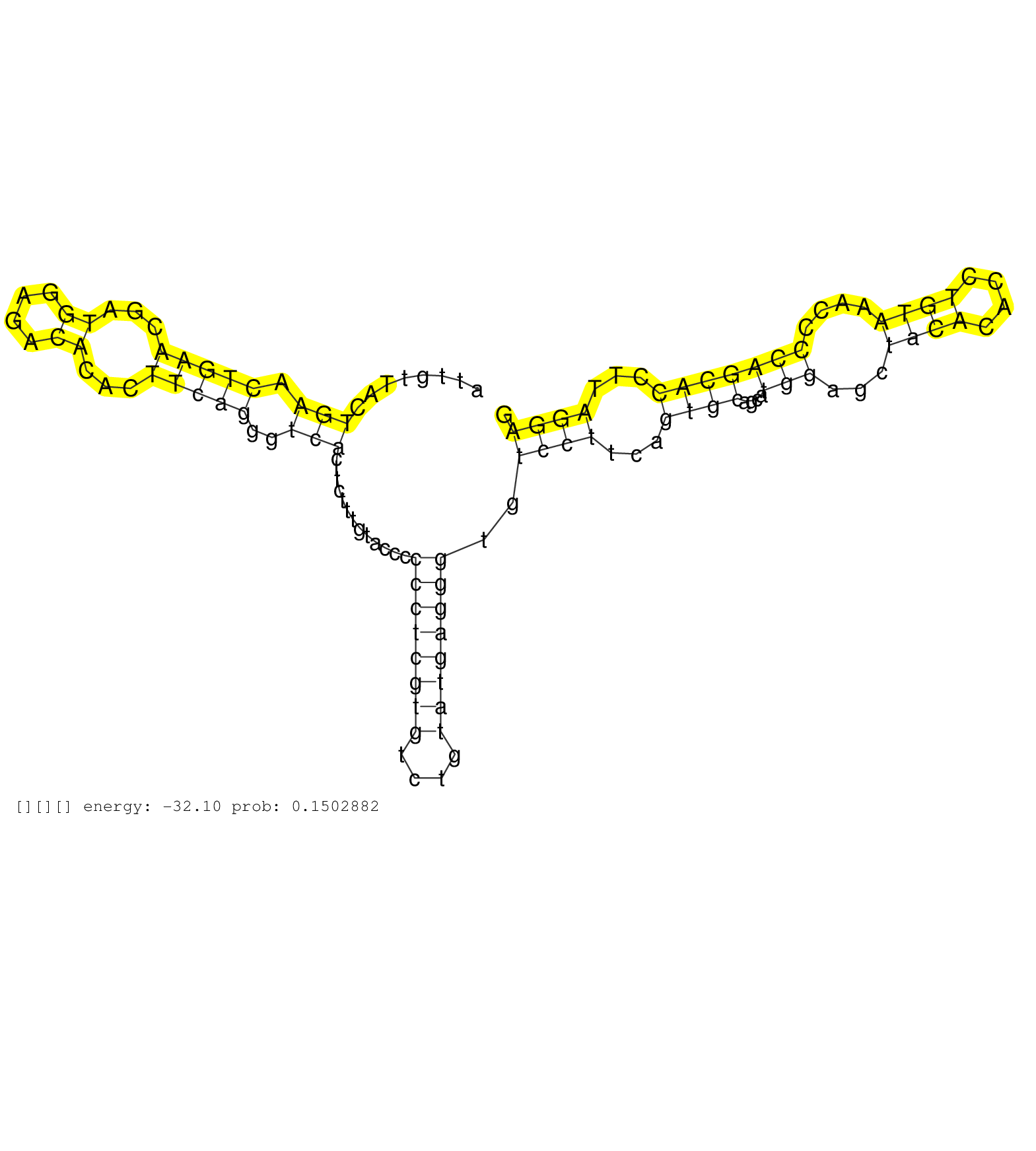

| Gene: Gtf2ird2 | ID: uc008zvd.1_intron_1_0_chr5_134667148_f.5p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(12) TESTES |
| CAGACGGCAGGATGGTGGTGACGTTCCTCATGTCTGCCCTGGAGTCCATGGTGAGACGCTGTGGCTCTGCCGATTGTTACTGAACTGAACGATGGAGACACACTTCAGGGTCACTTCTTTGTACCCCCCTCGTGTCTGTATGAGGGTGTCCTTCAGTGCAGCATGGAGCTACACACCTGTAAACCCCAGCACCTTAGGAGGTAGATAAAGAGGGCCAGGAGTTCAAGGTCATTAGAGGCATAAGGGCACT ................................................................................(((.(((((...((....))...)))))..))).............((((((((....))))))))..((((...((((....(((...((((....))))....)))))))...))))................................................... ........................................................................73.............................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................TACTGAACTGAACGATGGAGACACACTT................................................................................................................................................. | 28 | 1 | 7.00 | 7.00 | 4.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................TACTGAACTGAACGATGGAGACACACT.................................................................................................................................................. | 27 | 1 | 5.00 | 5.00 | - | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................TGAACTGAACGATGGAGACACACTTC................................................................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................TTGTTACTGAACTGAACGATGGAGAC....................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................TGTTACTGAACTGAACGATGGAGACAC..................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................CGATTGTTACTGAACTGAACGATGGAG......................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................................TAAAGAGGGCCAGGAGTTCAAGGTCAT.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................................TTAGGAGGTAGATAAAaaaa..................................... | 20 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................CTGTATGAGGGTGgct................................................................................................... | 16 | gct | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................ACTGAACTGAACGATGGAGACACACTT................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TACTGAACTGAACGATGGAGACACAC................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................CGATTGTTACTGAACTGAACGATGGA.......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................................GTAGATAAAGAGGGCCAGGAGTTCA......................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................................TAGGAGGTAGATAAAGAGGGCCAGGAG............................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................AGGAGGTAGATAAAGAcagt................................... | 20 | cagt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................AACTGAACGATGGAGACACACTTCAG.............................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................................................CACACCTGTAAACCCCAGCACCTTAGGAG.................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................TTACTGAACTGAACGATGGAGACACACTT................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| CAGACGGCAGGATGGTGGTGACGTTCCTCATGTCTGCCCTGGAGTCCATGGTGAGACGCTGTGGCTCTGCCGATTGTTACTGAACTGAACGATGGAGACACACTTCAGGGTCACTTCTTTGTACCCCCCTCGTGTCTGTATGAGGGTGTCCTTCAGTGCAGCATGGAGCTACACACCTGTAAACCCCAGCACCTTAGGAGGTAGATAAAGAGGGCCAGGAGTTCAAGGTCATTAGAGGCATAAGGGCACT ................................................................................(((.(((((...((....))...)))))..))).............((((((((....))))))))..((((...((((....(((...((((....))))....)))))))...))))................................................... ........................................................................73.............................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|