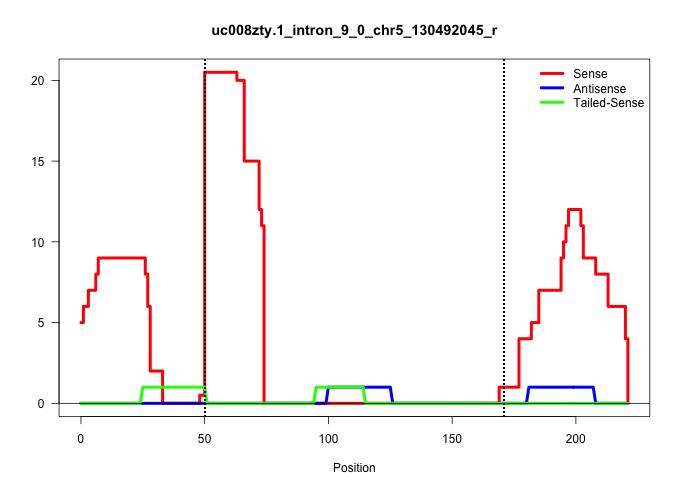
| Gene: Asl | ID: uc008zty.1_intron_9_0_chr5_130492045_r | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| ACACACTTGCAGAGAGCTCAGCCCATTCGCTGGAGCCACTGGATCCTGAGGTGAGTCGTGCGGAGCAGGGGCGGTAGGTAGGAAGCGGCCCTGACCAGGGGCTTTGATGGAAGAGGAGGCTCGCTAGTGTCCTACCACCAACCATCCACCCCTATTCCCTATTTCCCACAGCCACGCTGTTGCGCTGACACGAGATTCAGAGAGACTCTTGGAGGTGCAGA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTCGTGCGGAGCAGGGGCGG................................................................................................................................................... | 24 | 1 | 11.00 | 11.00 | 8.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTCGTGCGGAGC........................................................................................................................................................... | 16 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTCGTGCGGAGCAGGGGC..................................................................................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TGACACGAGATTCAGAGAGACTCTTGGA........ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................................TCAGAGAGACTCTTGGAGGTGCAGA | 25 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTTGCGCTGACACGAGATTCAGAGA.................. | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......TGCAGAGAGCTCAGCCCATTCGCTGG............................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CGCTGACACGAGATTCAGAGAGACTC............. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TTCAGAGAGACTCTTGGAGGTGCAGA | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGAGAGACTCTTGGAGGTGCAG. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................................................ATTCAGAGAGACTCTTGGAGGTGCAGA | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................AGCCACGCTGTTGCGCTGACACGAGAT......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .CACACTTGCAGAGAGCTCAGCCCATTC................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTCGTGCGGAGCAGGGGCG.................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................ATTCAGAGAGACTCTTGGAGGTGCAG. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...CACTTGCAGAGAGCTCAGCCCATTC................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTTGCGCTGACACGAGATTCAGAG................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................TTCGCTGGAGCCACTGGATCCTGAGc.......................................................................................................................................................................... | 26 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......TTGCAGAGAGCTCAGCCCATTCGCTGG............................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................CAGGGGCTTTGATGGAcgca.......................................................................................................... | 20 | cgca | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AGGTGAGTCGTGCGG.............................................................................................................................................................. | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ACACACTTGCAGAGAGCTCAGCCCATTCGCTGGAGCCACTGGATCCTGAGGTGAGTCGTGCGGAGCAGGGGCGGTAGGTAGGAAGCGGCCCTGACCAGGGGCTTTGATGGAAGAGGAGGCTCGCTAGTGTCCTACCACCAACCATCCACCCCTATTCCCTATTTCCCACAGCCACGCTGTTGCGCTGACACGAGATTCAGAGAGACTCTTGGAGGTGCAGA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................GCGCTGACACGAGATTCAGAGAGACTC............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............CTCAGCCCATTCGCat............................................................................................................................................................................................... | 16 | at | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................GCTTTGATGGAAGAGGAGGCTCGCTA............................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................CGCTGGAGCCACTGGATg................................................................................................................................................................................. | 18 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |