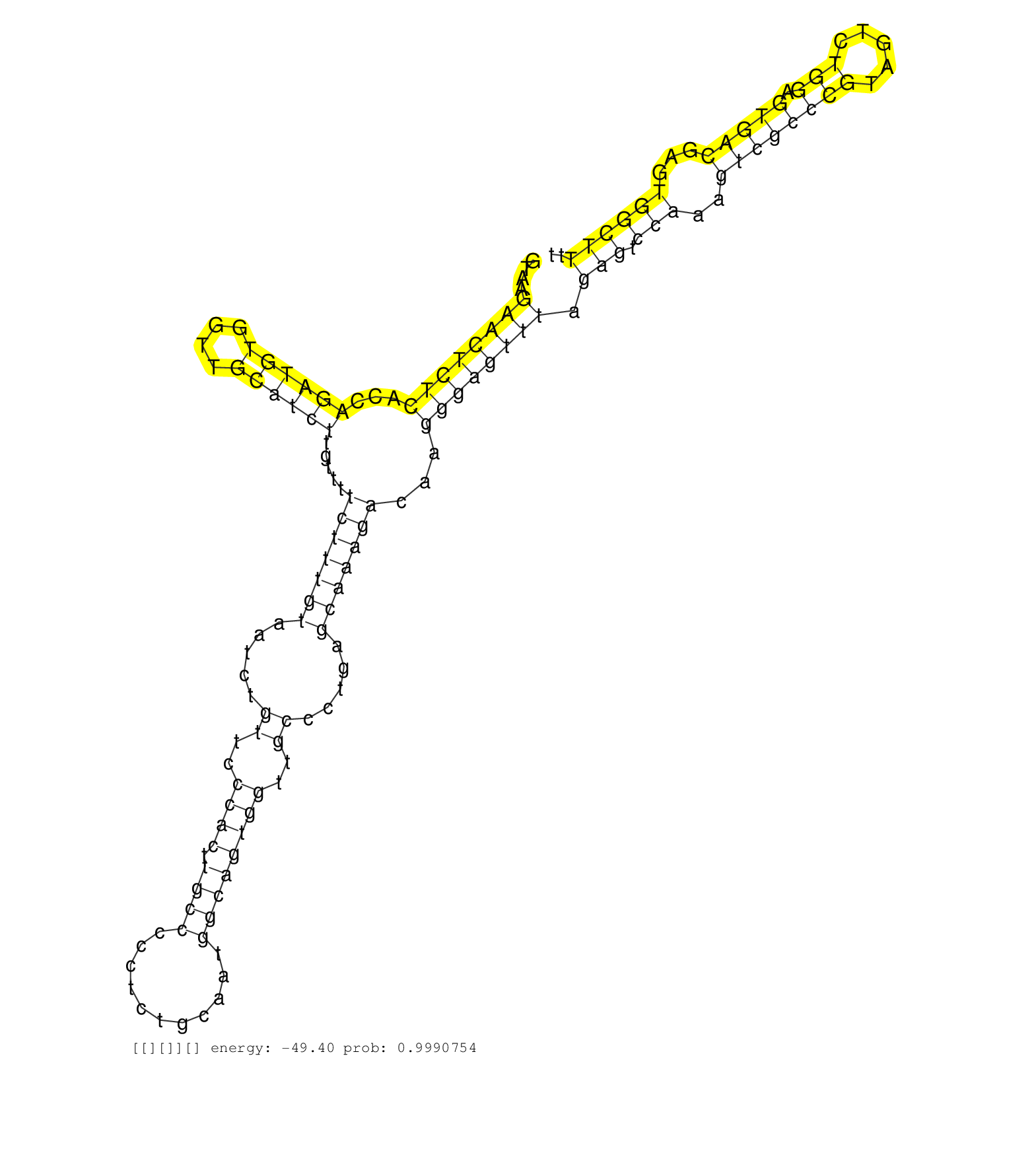

| Gene: Ccdc92 | ID: uc008zqr.1_intron_0_0_chr5_125316662_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(5) OTHER.mut |
(7) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(28) TESTES |
| TTTTGTAGATCTAACATATGAGCTGACACTCAAAAGTTTCGAACTGACAGGTAAGAACTCTCACCAGATGTGGTTGCATCTTGTTTTCTTTGTAATCTGTTCCCACTTGCCCCCTCTGCAATGGCAGTGGTTGCCCTGAGCAAAGACAAGGGAGTTTAGAGTCCAAAGTCGCCCGTAGTCTGGAGTGACGAGTGGCTTTTTCACTTCCCATTAATATATATGCCTAGAATGCCTATTGCATTTTCCAGAA ......................................................((((((((...((((((....)))))).....(((((((.....((..((((.((((...........))))))))..)).....)))))))...)))))))).(((.(((..((((((((.....))).)))))...)))))).................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGAACTCTCACCAGATGTGGTTGC............................................................................................................................................................................. | 27 | 1 | 17.00 | 17.00 | 4.00 | - | 1.00 | 1.00 | 3.00 | 2.00 | - | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 |
| .............................TCAAAAGTTTCGAACTGACAGGagac................................................................................................................................................................................................... | 26 | agac | 14.00 | 0.00 | 6.00 | - | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................AGCTGACACTCAAAAGTTTCGAACTGAC.......................................................................................................................................................................................................... | 28 | 1 | 8.00 | 8.00 | - | 2.00 | - | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACTCTCACCAGATGTGGTTGCA............................................................................................................................................................................ | 28 | 1 | 5.00 | 5.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACTCTCACCAGATGTGGTTGCAT........................................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................ATGAGCTGACACTCAAAAGT..................................................................................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGACACTCAAAAGTTTCGAACTGACAGG....................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................TCAAAAGTTTCGAACTGACAGGaga.................................................................................................................................................................................................... | 25 | aga | 2.00 | 0.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGCTGACACTCAAAAGTTTCGAACTGACA......................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........ATCTAACATATGAGCTGACACTCAAAA....................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGCTGACACTCAAAAGTTTCGAACTGACAGa....................................................................................................................................................................................................... | 31 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............ACATATGAGCTGACACTCAAAAGTTTCGAAC.............................................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CAAAAGTTTCGAACTGACAGGagac................................................................................................................................................................................................... | 25 | agac | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGCTGACACTCAAAAGTTTCGAACTGAa.......................................................................................................................................................................................................... | 28 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TATGAGCTGACACTCAAAAGT..................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................ACACTCAAAAGTTTCGAACTGACAGagc..................................................................................................................................................................................................... | 28 | agc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................CGTAGTCTGGAGTGACGAGTGGCTT.................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAAGAACTCTCACCAGATGTGGTTG.............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGCTGACACTCAAAAGTTTCGAACTG............................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GATCTAACATATGAGCTGACACTCAAAAG...................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CAAAAGTTTCGAACTGACAGGaga.................................................................................................................................................................................................... | 24 | aga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TTTCGAACTGACAGGaga.................................................................................................................................................................................................... | 18 | aga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGCTGACACTCAAAAGTTTCGAACTGACAGag...................................................................................................................................................................................................... | 32 | ag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGCTGACACTCAAAAGTTTCGAACT............................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............ATATGAGCTGACACTCAAAAGTTTCGAAC.............................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGACACTCAAAAGTTTCGAACTGACAGag...................................................................................................................................................................................................... | 29 | ag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................AGCTGACACTCAAAAGTTTCGAACTGACtga....................................................................................................................................................................................................... | 31 | tga | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GATCTAACATATGAGCTGACACTCAAAA....................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TCAAAAGTTTCGAACTGACAGGag..................................................................................................................................................................................................... | 24 | ag | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TATGAGCTGACACTCAAAAGTTTCGAACTGA........................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACTCTCACCAGATGTGGTTGCATC.......................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTTTGTAGATCTAACATATGAGCTGACACTCAAAAGTTTCGAACTGACAGGTAAGAACTCTCACCAGATGTGGTTGCATCTTGTTTTCTTTGTAATCTGTTCCCACTTGCCCCCTCTGCAATGGCAGTGGTTGCCCTGAGCAAAGACAAGGGAGTTTAGAGTCCAAAGTCGCCCGTAGTCTGGAGTGACGAGTGGCTTTTTCACTTCCCATTAATATATATGCCTAGAATGCCTATTGCATTTTCCAGAA ......................................................((((((((...((((((....)))))).....(((((((.....((..((((.((((...........))))))))..)).....)))))))...)))))))).(((.(((..((((((((.....))).)))))...)))))).................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|