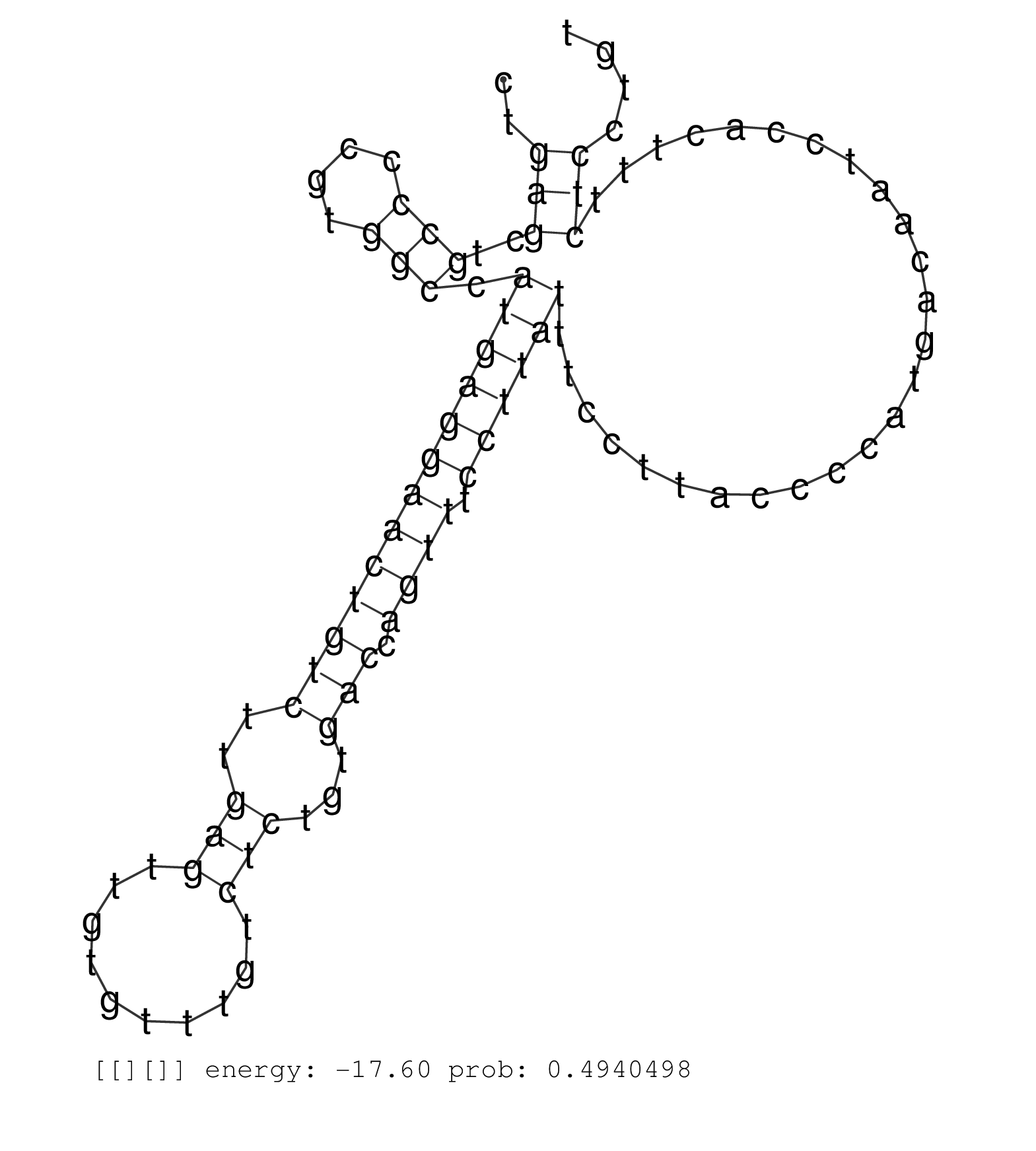

| Gene: Tctn2 | ID: uc008zqh.1_intron_10_0_chr5_125066556_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(7) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| GTTTTTAAGCCATCACAGTGGTGGTGAGAAGGAATTTTCTGGAAATCCAGGTAGGATGAATGACAGTGCTGCTCTTCTACACAGATCTTAGGGTGGCATCCTGAGCTGCCCCGTGGCCATGAGGAACTGTCTTGAGTTGTGTTTGTCTCTGTGACCAGTTTCCTTATTTCCTTACCCCATGACAATCCACTTTCTCCTGTGGCAGATTTATACTTTTTTTTTCTTTTTTTTGAGACAGGGTTTCTCTGTA ......................................................................................................(((..(((....))).(((((((((((((..(((..........)))...))).)))).))))))..........................)))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGATGAATGACAGTGCTGCTCTTC............................................................................................................................................................................. | 27 | 1 | 23.00 | 23.00 | 7.00 | 3.00 | 2.00 | - | 4.00 | 1.00 | 2.00 | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | - |
| ..................................................GTAGGATGAATGACAGTGCTGCTCTT.............................................................................................................................................................................. | 26 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TGAGAAGGAATTTTCTGGAAATCCAGGT...................................................................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................TGAGAAGGAATTTTCTGGAAATCCAGGTt..................................................................................................................................................................................................... | 29 | t | 2.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ..................................................GTAGGATGAATGACAGTGCTGCTCTTCTA........................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGATGAATGACAGTGCTGCTCTTCT............................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TGAGAAGGAATTTTCTGGAAATCCAGG....................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TGAGAAGGAATTTTCTGGAAATCCAGctt..................................................................................................................................................................................................... | 29 | ctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................AATTTTCTGGAAATCCAGG....................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| GTTTTTAAGCCATCACAGTGGTGGTGAGAAGGAATTTTCTGGAAATCCAGGTAGGATGAATGACAGTGCTGCTCTTCTACACAGATCTTAGGGTGGCATCCTGAGCTGCCCCGTGGCCATGAGGAACTGTCTTGAGTTGTGTTTGTCTCTGTGACCAGTTTCCTTATTTCCTTACCCCATGACAATCCACTTTCTCCTGTGGCAGATTTATACTTTTTTTTTCTTTTTTTTGAGACAGGGTTTCTCTGTA ......................................................................................................(((..(((....))).(((((((((((((..(((..........)))...))).)))).))))))..........................)))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................ACAGTGCTGCTCTTCTACACAGATCTTA................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CATGACAATCCACTTTCTCCTGTGGCA.............................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................GAAATCCAGGTAGGATtgt................................................................................................................................................................................................. | 19 | tgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................TGGCATCCTGAGCTGCta............................................................................................................................................. | 18 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |