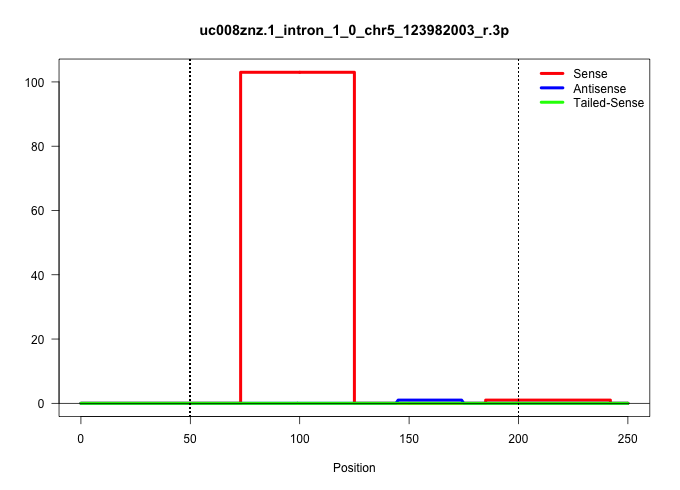
| Gene: Vps33a | ID: uc008znz.1_intron_1_0_chr5_123982003_r.3p | SPECIES: mm9 |
 |
|
(2) PIWI.ip |
(4) TESTES |
| TGGATTCTACAGGCAGCTGCATTCTTACATGCATACACACGCAAGCAGATCCATGCTGTACCCTTAAATAAAAGTAAACTTGAAAAAGTGCAGTCCTGGATGGTAAGGGTAAAGAGGTGGATTGGAGCAAGGCATATTTTGAGTTTCTGTTTCCCATCATATAACTTCAGTCCTGTATCTTTTTTTTTAACCATTGACAGAACCCCACAGACATATCCTATGTGTACAGCGGTTATGCTCCACTCAGCGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|
| .........................................................................GTAAACTTGAAAAAGTGCAGTCCTGGATGGTAAGGGTAAAGAGGTGGATTGG............................................................................................................................. | 52 | 1 | 103.00 | 103.00 | 103.00 | - | - | - |
| .........................................................................................................................................................................................TTTAACCATTGACAGAACCCCACAGACA..................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .....................................................................................................................................................................................................................TATCCTATGTGTACAGCGGTTATGCTCCA........ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| TGGATTCTACAGGCAGCTGCATTCTTACATGCATACACACGCAAGCAGATCCATGCTGTACCCTTAAATAAAAGTAAACTTGAAAAAGTGCAGTCCTGGATGGTAAGGGTAAAGAGGTGGATTGGAGCAAGGCATATTTTGAGTTTCTGTTTCCCATCATATAACTTCAGTCCTGTATCTTTTTTTTTAACCATTGACAGAACCCCACAGACATATCCTATGTGTACAGCGGTTATGCTCCACTCAGCGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................TCTGTTTCCCATCATATAACTTCAGTCCTG........................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |