
| Gene: Mlxip | ID: uc008znq.1_intron_6_0_chr5_123893975_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(15) TESTES |
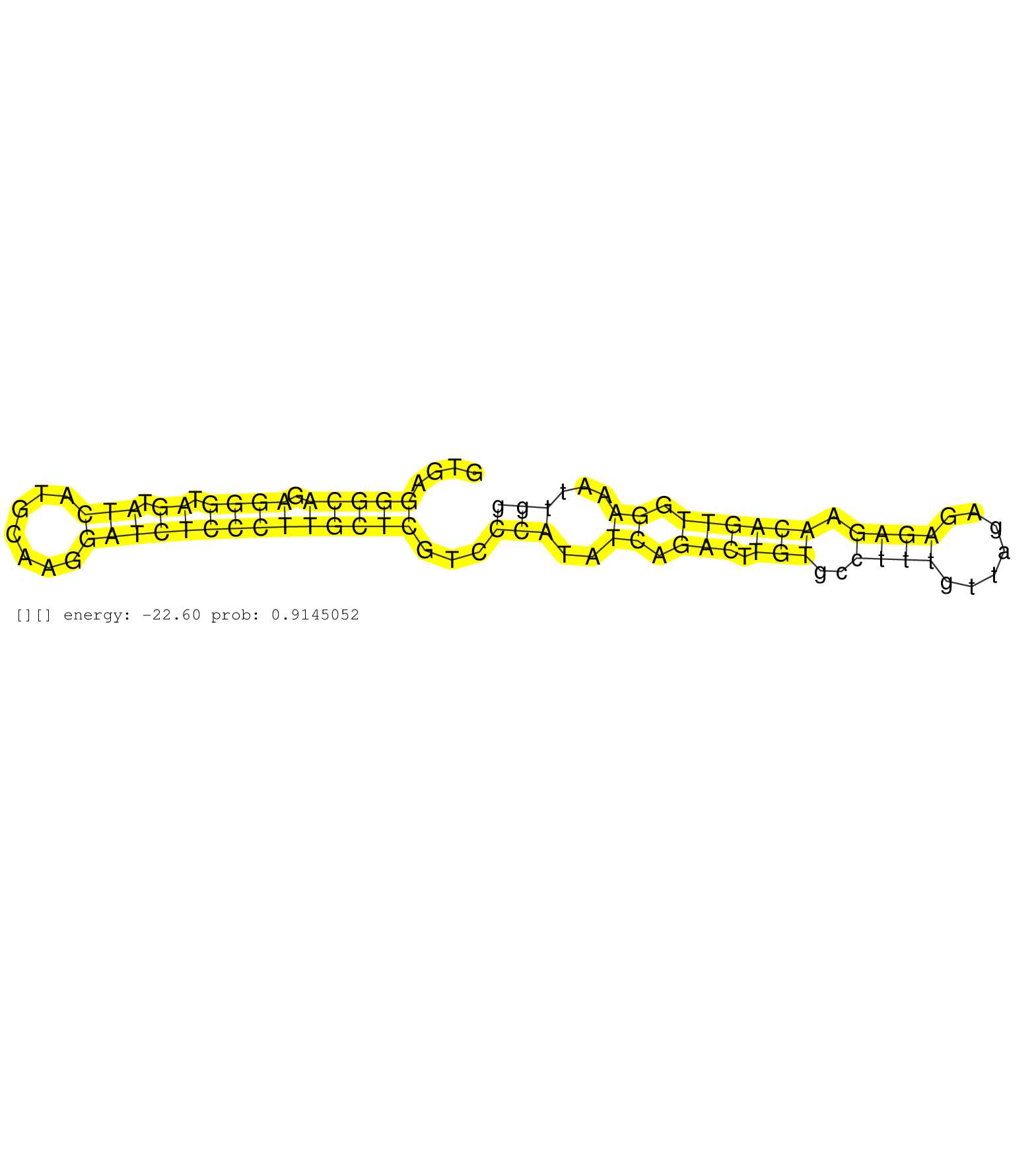
| CCCTTTGCAACCCAACCTGGACTTCATGGACACCTTTGAGCCTTTCCAGGGTGAGGGCAGAGGGTAGTATCATGCAAGGATCTCCCTTGCTCGTCCCATATCAGACTTGTGCCTTTGTTAGAGAGAGAACAGTTGGAAATTGGTGTAAAAGTGGGGTGGGGCTTATAAGTTTTTAAGACTGAAAATGATGTTGGTAGTAACTGCAAAGCCTGGGAAAGTTTCCTGGGACATTGGAAGGAAGTTGGCATCTCAGTGAGTGCTTTCTTCCTTGATTGTCAGCAGTTCCTCCACTAGACAGCAGAGGGCACTGTGACACAGCCACCACCCAGCAATGTTTCTGTATTTAGGTCAGACTCCCAGAGCTCTGCCTGGAAGCACTTGGTGTTTCAAGTGGTCCAATCAGAGCCCTCTTTCTACAAAACAAGTTATCTGGGGAGGCGGGGTTTGGGTCCAGACCCCCGTTTGTGTGTCATAATTCCAGAAAATGCTAAGTTCATGGAGTTCAGAAGACTCCCGTGTTCTTTTCCAGATCTCTTCTCCTCCAGCCGATCCATTTTTGGCTCCATGTTGCCTCCTCCT ......................................................(((((.((((.((.(((.......))))))))))))))...(((..((.(((.(((..((((.......)))).)))))).))...))).................................................................................................................................................................................................................................................................................................................................................................................................................................................... ..................................................51..........................................................................................143.................................................................................................................................................................................................................................................................................................................................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGGGCAGAGGGTAGTATCATGCAAGGATCTCCCTTGCTCGTCCCATATC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 52 | 1 | 40.00 | 40.00 | 40.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TTGCAACCCAACCTGGACTTCATGGAC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - |
| ....TTGCAACCCAACCTGGACTTCATGGACA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....TGCAACCCAACCTGGACTTCATGGACA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGCAACCCAACCTGGACTTCA......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................AGAGAGAACAGTTGGggc........................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 18 | ggc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................ATGTTGGTAGTAACTGC....................................................................................................................................................................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................TGGACACCTTTGAGCCTTTCCAGGatc.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 27 | atc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................TGTATTTAGGTCAGACTCCCAGAGCTCTGC................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................TAGAGAGAGAACAGTTGGAAATTGGT................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...TTTGCAACCCAACCTGGACTTCATGGA..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................GAAAATGATGTTGGgagc............................................................................................................................................................................................................................................................................................................................................................................................. | 18 | gagc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................TCATGGACACCTTTGAGCCTTTC..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TTTGCAACCCAACCTGGACTTCAcgga..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | cgga | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ATGGAGTTCAGAAGACTCCCG............................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................CTTCATGGACACCTTTGAGCCTTTCCAGGa................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 30 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................TAGAGAGAGAACAGTTGGAAATTGGTGT................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................................................................TTCCTTGATTGTCAGCAGT........................................................................................................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| CCCTTTGCAACCCAACCTGGACTTCATGGACACCTTTGAGCCTTTCCAGGGTGAGGGCAGAGGGTAGTATCATGCAAGGATCTCCCTTGCTCGTCCCATATCAGACTTGTGCCTTTGTTAGAGAGAGAACAGTTGGAAATTGGTGTAAAAGTGGGGTGGGGCTTATAAGTTTTTAAGACTGAAAATGATGTTGGTAGTAACTGCAAAGCCTGGGAAAGTTTCCTGGGACATTGGAAGGAAGTTGGCATCTCAGTGAGTGCTTTCTTCCTTGATTGTCAGCAGTTCCTCCACTAGACAGCAGAGGGCACTGTGACACAGCCACCACCCAGCAATGTTTCTGTATTTAGGTCAGACTCCCAGAGCTCTGCCTGGAAGCACTTGGTGTTTCAAGTGGTCCAATCAGAGCCCTCTTTCTACAAAACAAGTTATCTGGGGAGGCGGGGTTTGGGTCCAGACCCCCGTTTGTGTGTCATAATTCCAGAAAATGCTAAGTTCATGGAGTTCAGAAGACTCCCGTGTTCTTTTCCAGATCTCTTCTCCTCCAGCCGATCCATTTTTGGCTCCATGTTGCCTCCTCCT ......................................................(((((.((((.((.(((.......))))))))))))))...(((..((.(((.(((..((((.......)))).)))))).))...))).................................................................................................................................................................................................................................................................................................................................................................................................................................................... ..................................................51..........................................................................................143.................................................................................................................................................................................................................................................................................................................................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................................................AGTGAGTGCTTTCTTCCTTGATTGTCA............................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |