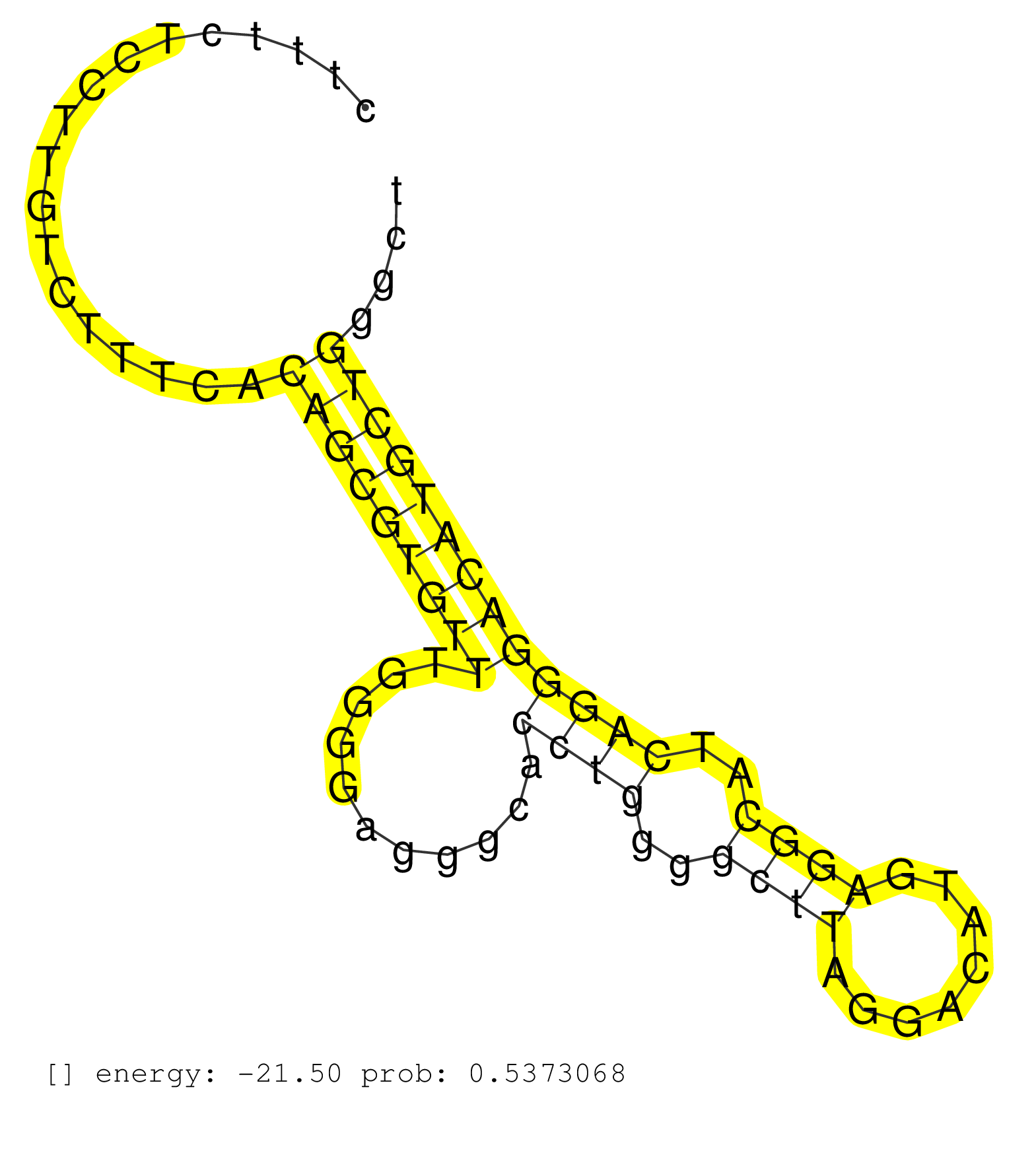

| Gene: mKIAA0787 | ID: uc008zmb.1_intron_6_0_chr5_123196459_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| AATTGCTATAGGCGGTTGATGTGTTGAGAATCGTGTAAAGGTCCTTGCACACACTTTCTCCTTGTCTTTCACAGCGTGTTTGGGGAGGGCACCTGGGGCTTAGGACATGAGGCATCAGGGACATGCTGGGCTGCATGGCCAGGTGGAGATTTTCACGGGGTCTCCCTGCCCCGTCTCCTCCTGGTCTCCTCTGTGTCTAGTGCATTACCAGAAGATCATCCATCGGGACATCAAACCTTCCAACCTCCTG .......................................................................(((((((((...........((((..((((........))))..))))))))))))).......................................................................................................................... .....................................................54............................................................................132.................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................TAGGACATGAGGCATCAGGGACATGCTG.......................................................................................................................... | 28 | 1 | 33.00 | 33.00 | 8.00 | 3.00 | 5.00 | 5.00 | - | 5.00 | 1.00 | - | 1.00 | - | 3.00 | - | - | - | - | - | - | 1.00 | 1.00 | - |
| ....................................................................................................TAGGACATGAGGCATCAGGGACATGCT........................................................................................................................... | 27 | 1 | 20.00 | 20.00 | 2.00 | 3.00 | 6.00 | 1.00 | - | 1.00 | - | 1.00 | 2.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | 1.00 |
| ....................................................................................................TAGGACATGAGGCATCAGGGACATGC............................................................................................................................ | 26 | 1 | 15.00 | 15.00 | 3.00 | 2.00 | 1.00 | 2.00 | 3.00 | - | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................TAGGACATGAGGCATCAGGGACATG............................................................................................................................. | 25 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................GCTTAGGACATGAGGCATCAGG................................................................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................TAGGACATGAGGCATCAGGGACAT.............................................................................................................................. | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GGGGCTTAGGACATGAGGCA........................................................................................................................................ | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TAGGACATGAGGCATCAGGGACATGt............................................................................................................................ | 26 | t | 2.00 | 3.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TAGGACATGAGGCATCAGGGACATGCTGG......................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TTGTCTTTCACAGCGTGTTTGGGGgg................................................................................................................................................................... | 26 | gg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................TTAGGACATGAGGCATCAGGGAC................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................TAGGACATGAGGCATCAGGGACATGCTt.......................................................................................................................... | 28 | t | 1.00 | 20.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TCCTTGTCTTTCACAGCGTGTTTGGGG..................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TGAGAATCGTGTAAAGGTCCTTGCAC........................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TAGGACATGAGGCATCAGGGACA............................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................CTTTCACAGCGTGTTTGGGGAGGtc................................................................................................................................................................ | 25 | tc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GGCTTAGGACATGAGGCATCAGGGACAT.............................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TAGGACATGAGGCATCAGGGACATGCTGGG........................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TGCATTACCAGAAGATCATCCATCGGGAC..................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TTAGGACATGAGGCATCAGGGACATGCT........................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................GAAGATCATCCATCGGGACATCAAAC.............. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TTAGGACATGAGGCATCAGGGACATGC............................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TTAGGACATGAGGCATCAGGGACAT.............................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GGCTTAGGACATGAGGCATCAGGGAC................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TGCATTACCAGAAGATCATCCATCGG........................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TAGGACATGAGGCATCAGGGACATGCTGt......................................................................................................................... | 29 | t | 1.00 | 33.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................GCTTAGGACATGAGGCATCAGGGACAT.............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TTTCTCCTTGTCTTTCACAGCGTGTT.......................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| AATTGCTATAGGCGGTTGATGTGTTGAGAATCGTGTAAAGGTCCTTGCACACACTTTCTCCTTGTCTTTCACAGCGTGTTTGGGGAGGGCACCTGGGGCTTAGGACATGAGGCATCAGGGACATGCTGGGCTGCATGGCCAGGTGGAGATTTTCACGGGGTCTCCCTGCCCCGTCTCCTCCTGGTCTCCTCTGTGTCTAGTGCATTACCAGAAGATCATCCATCGGGACATCAAACCTTCCAACCTCCTG .......................................................................(((((((((...........((((..((((........))))..))))))))))))).......................................................................................................................... .....................................................54............................................................................132.................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................TGGTCTCCTCTGTGtgct....................................................... | 18 | tgct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |