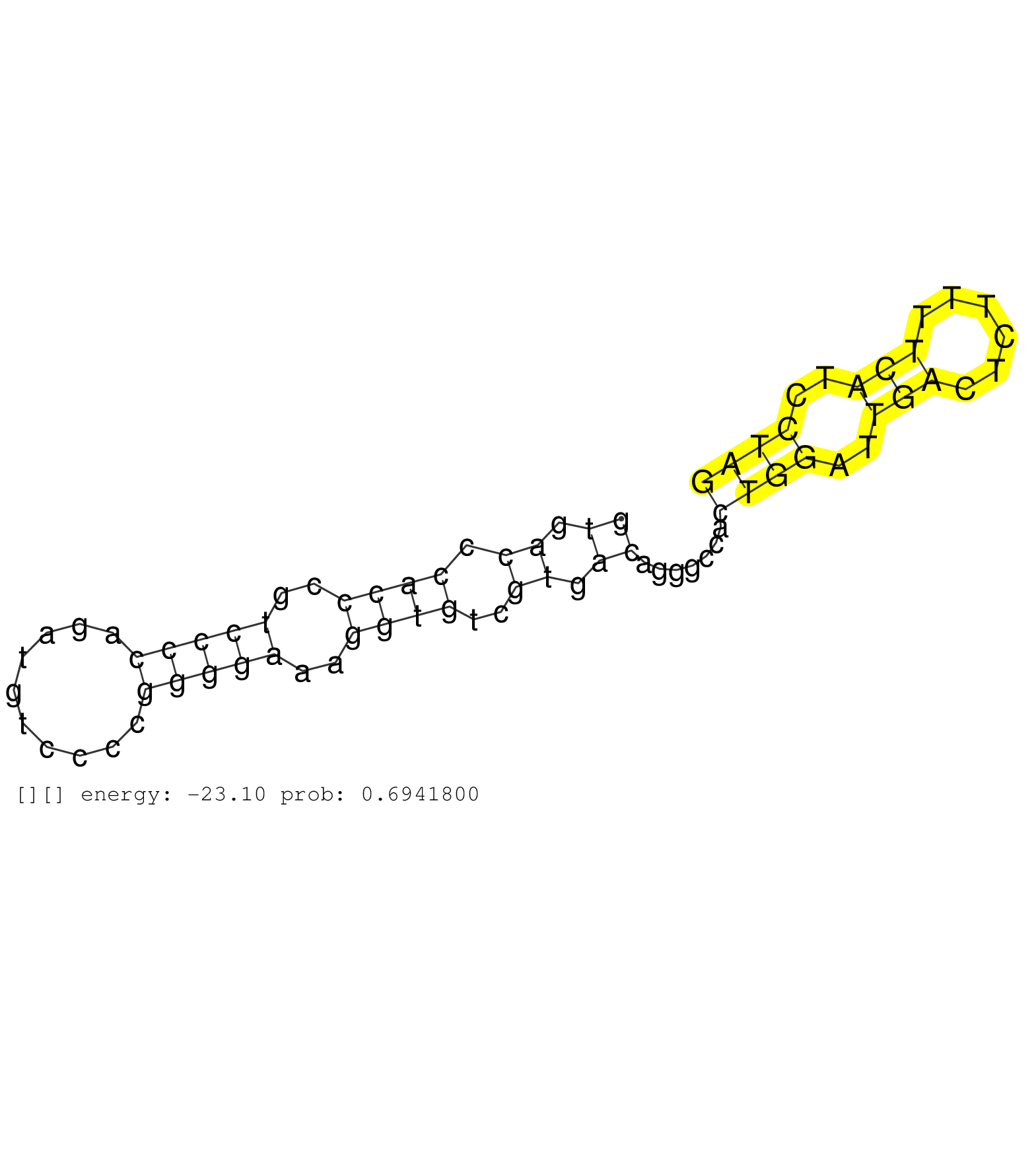
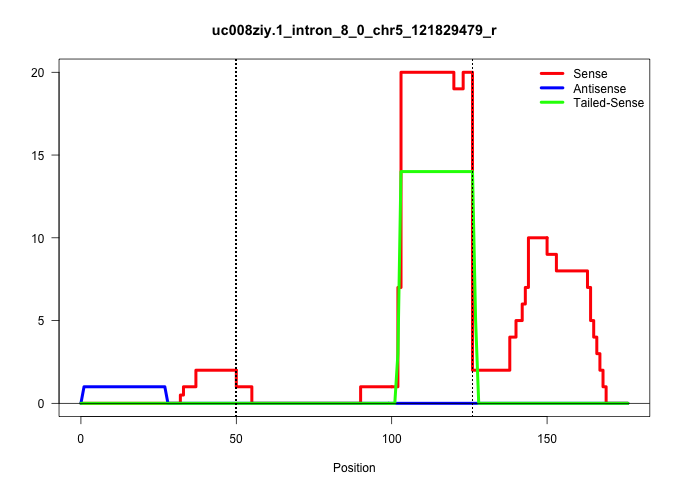
| Gene: Trafd1 | ID: uc008ziy.1_intron_8_0_chr5_121829479_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| CGTTCCCCAAATCTGACATGGACATTCACATGGCTGCAGAGCACTGTCAGGTGACCCACCCGTCCCCAGATGTCCCCGGGGAAAGGTGTCGTGACAGGGCCACTGGATTGACTCTTTTCATCCTAGGTGACCTGTAAATGCAACAAGAAGTTGGAGAAGAGGCAGTTAAAGCAGCA ..................................................((.((.((((..(((((..........)))))..))))..)).)).......((((..(((......)))..)))).................................................. ..................................................51.........................................................................126................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT1() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................TGGATTGACTCTTTTCATCCTAG.................................................. | 23 | 1 | 16.00 | 16.00 | 8.00 | 4.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGGATTGACTCTTTTCATCCTAGa................................................. | 24 | a | 10.00 | 16.00 | 4.00 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CTGGATTGACTCTTTTCATCCTAG.................................................. | 24 | 1 | 7.00 | 7.00 | 1.00 | 1.00 | 1.00 | - | - | - | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - |
| .............................ATGGCTGCAGAGCACTGTCAG.............................................................................................................................. | 21 | 1 | 5.00 | 5.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGGATTGACTCTTTTCATCCTAGaa................................................ | 25 | aa | 4.00 | 16.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TCTGACATGGACATTCACATGGCTGCAGA........................................................................................................................................ | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TGCAACAAGAAGTTGGAGAAGAGGCA............ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................................CTGGATTGACTCTTTTCATCCTAGa................................................. | 25 | a | 2.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................TGGACATTCACATGGCTGC........................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - |
| ..............................................................................................................................................ACAAGAAGTTGGAGAAGAGGCAGTTA........ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................GTGACCTGTAAATGCAACAAGAAGTTG....................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AAGAAGTTGGAGAAGAGGCAGTT......... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AAGAAGTTGGAGAAGAGGCAGT.......... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AAGAAGTTGGAGAAGAGGC............. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CAAGAAGTTGGAGAAGAGGCAGTTAA....... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................GTGACAGGGCCACTGGATTGACTCTTTTCA........................................................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................AGAGCACTGTCAGGTGAC......................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TAGGTGACCTGTAAATGCAACAAGAAG.......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................CTGGATTGACTCTTTTCATCCTAGaa................................................ | 26 | aa | 1.00 | 7.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CAACAAGAAGTTGGAGAAGAGGCAG........... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................CTGCAGAGCACTGTCAG.............................................................................................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ................................GCTGCAGAGCACTGTCAG.............................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| CGTTCCCCAAATCTGACATGGACATTCACATGGCTGCAGAGCACTGTCAGGTGACCCACCCGTCCCCAGATGTCCCCGGGGAAAGGTGTCGTGACAGGGCCACTGGATTGACTCTTTTCATCCTAGGTGACCTGTAAATGCAACAAGAAGTTGGAGAAGAGGCAGTTAAAGCAGCA ..................................................((.((.((((..(((((..........)))))..))))..)).)).......((((..(((......)))..)))).................................................. ..................................................51.........................................................................126................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT1() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .GTTCCCCAAATCTGACATGGACATTCA.................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |