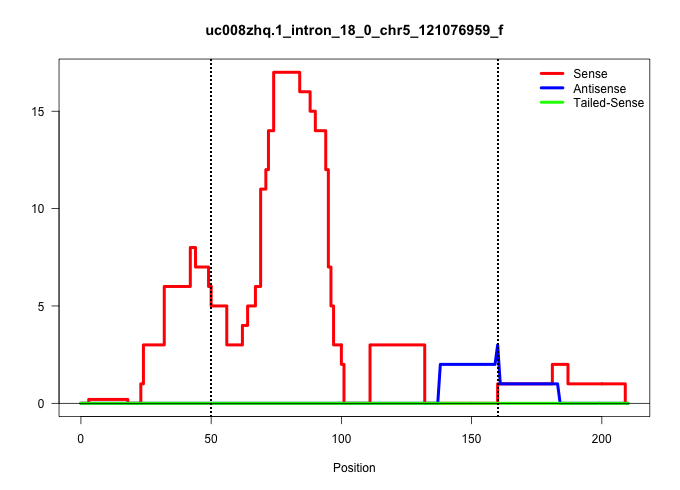
| Gene: Ddx54 | ID: uc008zhq.1_intron_18_0_chr5_121076959_f | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| CCAGCGAGGCCCTGGGCCCCGCAGAGGTGGAAAGCGAGGTCGTAGTCAAGGTGGGCAGGGCAGGTGGTATGGTGTGAACAGGAAGAAAGCCAGGCATGGTGAGGCACAGGGATGAGCACGGGCCCAAGGCTGAGCTCCCCCCAAATCCCTGTGTTGACAGGCACATCCCAGCCCCGAGCTTCCAGTGTACCCGCAGGCCGCATGCGCTCG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................TGGTGTGAACAGGAAGAAAGCCAGGC................................................................................................................... | 26 | 1 | 5.00 | 5.00 | 2.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................ATGAGCACGGGCCCAAGGCTG.............................................................................. | 21 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................AGCGAGGTCGTAGTCAAGGTGGGC.......................................................................................................................................................... | 24 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGAACAGGAAGAAAGCCAGGCATGGTG............................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................TGTGAACAGGAAGAAAGCCAGGCAT................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................TAGTCAAGGTGGGCAGGGCAGGTGGTA............................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................GAGGTGGAAAGCGAGGTCGTA...................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................AGGTGGAAAGCGAGGTCGTAGTCAAG................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................GTGTGAACAGGAAGAAAGCCAGGCA.................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................TGAACAGGAAGAAAGCCAGGCATGGT.............................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CCAGTGTACCCGCAGGCCGCATGCGCTC. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................GCACATCCCAGCCCCGAGCTTCCAGTG....................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................AGGGCAGGTGGTATGGTGTGAACAGGAA.............................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................TGGTATGGTGTGAACAGGAAGAAAGC........................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GGTGGTATGGTGTGAACAGGAAGAAA.......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AGGTGGAAAGCGAGGTCGTAGTCAA................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................TGGTGTGAACAGGAAGAAAGCCAGG.................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TATGGTGTGAACAGGAAGAAAGCCAGG.................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TGGTGTGAACAGGAAGAAAGCCAGGCA.................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...GCGAGGCCCTGGGCC................................................................................................................................................................................................ | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - |
| CCAGCGAGGCCCTGGGCCCCGCAGAGGTGGAAAGCGAGGTCGTAGTCAAGGTGGGCAGGGCAGGTGGTATGGTGTGAACAGGAAGAAAGCCAGGCATGGTGAGGCACAGGGATGAGCACGGGCCCAAGGCTGAGCTCCCCCCAAATCCCTGTGTTGACAGGCACATCCCAGCCCCGAGCTTCCAGTGTACCCGCAGGCCGCATGCGCTCG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................CCCCAAATCCCTGTGTTGACAGG................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................GCACATCCCAGCCCCGAGCTTCCA.......................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................TCCCTGTGTTGACAggat................................................... | 18 | ggat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |