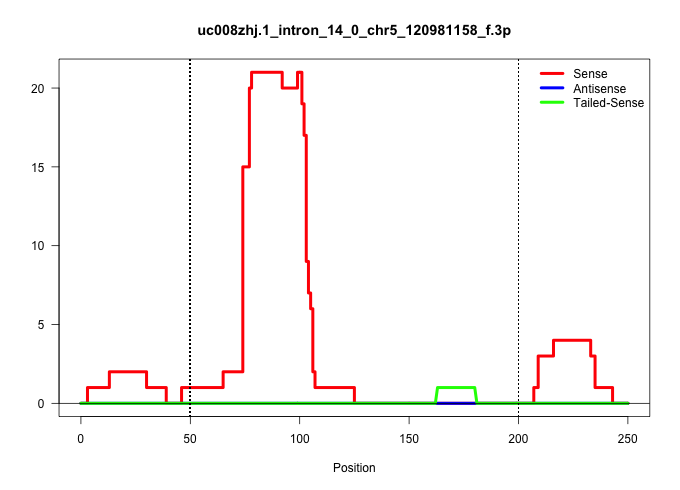
| Gene: Slc24a6 | ID: uc008zhj.1_intron_14_0_chr5_120981158_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| CCATGGGAATGCATGTCTATCGCTGCTAACACGGGAGGCCAGGAAGAAAAGCCCAAACAGGCGGATGTGGACGCTGATAGAGACAGTACCACGTGGCAGACCCCATAGCTGTGTACCAAGGACCCAACACTCCCTTCATGGTGCACATGGCCCAGAAGGCTCCCAGGGTGGGGACTAAACAACTCCTGTTCTCCCCACAGCTGGAGCCAGACGGATTACTGGTGTGGGTGCTGGCCAGTGCCCTGGGCCT ..................................................(((......)))..((((((..((((.......)))).))))))............................................................................................................................................................ ..................................................51......................................................107............................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................TGATAGAGACAGTACCACGTGGCAGACCC................................................................................................................................................... | 29 | 1 | 8.00 | 8.00 | 4.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - |
| .............................................................................TAGAGACAGTACCACGTGGCAGACCCCAT................................................................................................................................................ | 29 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGATAGAGACAGTACCACGTGGCAGACCCC.................................................................................................................................................. | 30 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ..........................................................................TGATAGAGACAGTACCACGTGGCAGACC.................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................................................................................................GACGGATTACTGGTGTGGGTGCTGGC............... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| ..........................................................................TGATAGAGACAGTACCACGTGGCAGAC..................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................AGAGACAGTACCACGTGGCAGACCCCAT................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................AAAAGCCCAAACAGGCGGATGTGGACGC................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............TGTCTATCGCTGCTAACACGGGAGGC................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................CAGGGTGGGGACTAcata..................................................................... | 18 | cata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...TGGGAATGCATGTCTATCGCTGCTAAC............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................TAGAGACAGTACCACGTGGCAGACCCCATA............................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................TAGCTGTGTACCAAGGACCCAACAC........................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................CAGACGGATTACTGGTGTGGGTGCTG................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................................................TACTGGTGTGGGTGCTGGCCAGTGCCC....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................TAGAGACAGTACCACGTGGCAGACCCCA................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................TGTGGACGCTGATAGAGACAGTACCAC.............................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................ACCCCATAGCTGTGTACCAAGGACCC............................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| CCATGGGAATGCATGTCTATCGCTGCTAACACGGGAGGCCAGGAAGAAAAGCCCAAACAGGCGGATGTGGACGCTGATAGAGACAGTACCACGTGGCAGACCCCATAGCTGTGTACCAAGGACCCAACACTCCCTTCATGGTGCACATGGCCCAGAAGGCTCCCAGGGTGGGGACTAAACAACTCCTGTTCTCCCCACAGCTGGAGCCAGACGGATTACTGGTGTGGGTGCTGGCCAGTGCCCTGGGCCT ..................................................(((......)))..((((((..((((.......)))).))))))............................................................................................................................................................ ..................................................51......................................................107............................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|