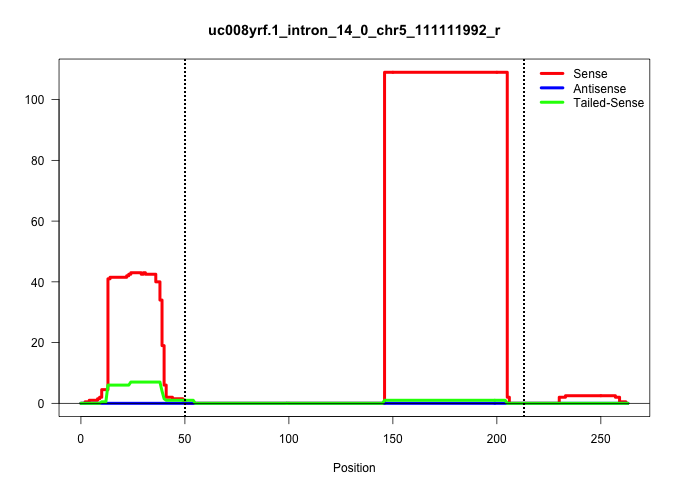
| Gene: Ep400 | ID: uc008yrf.1_intron_14_0_chr5_111111992_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(20) TESTES |
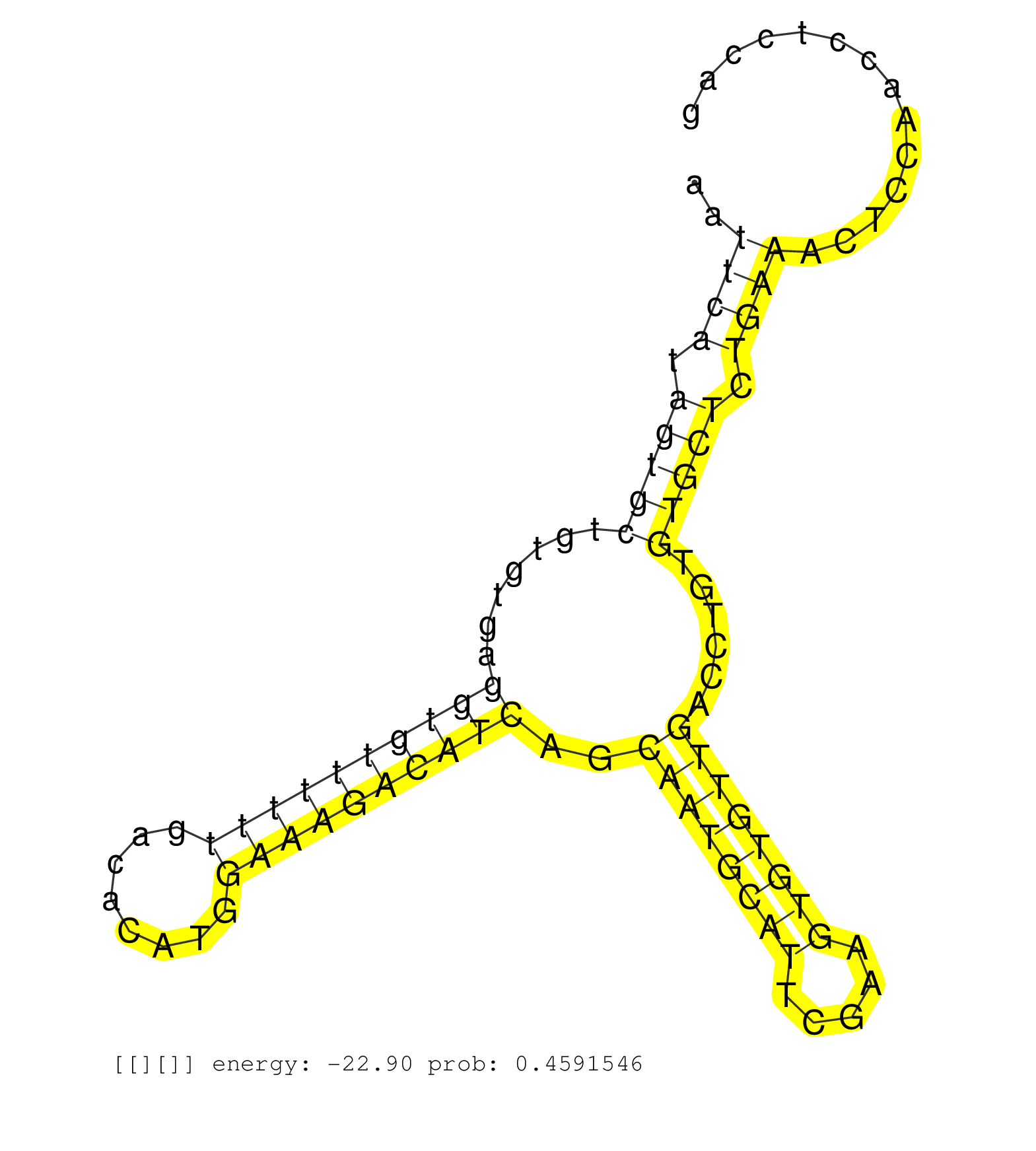
| CCCAGGACAGTCCTGACTGGCTCATTGGCGAGGACTGGGCCTTGCTACAGGTGAGTGGCTGTGTTATCTTTGAGTTGAGCATTCCTGTGGTTTGTTTGCTCCCCCACATTGGCAATTCATAGTGCTGTGTGAGGTGTTTTTTGACACATGGAAAGACATCAGCAATGCATTCGAAGTGTGTTGACCTGTGTGCTCTGAAACTCCAACCTCCAGGCTGTAAAGCAGTTGCTTGAGCTGCCCTTGAACCTCACGATTGTGTCACC ...................................................................................................................((((.(((((.......((((((((((........))))))))))..((((((((.....))))))))......))))).))))................................................................ .................................................................................................................114................................................................................................213................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................CATGGAAAGACATCAGCAATGCATTCGAAGTGTGTTGACCTGTGTGCTCTGA................................................................. | 52 | 1 | 403.00 | 403.00 | 403.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TGACTGGCTCATTGGCGAGGACTGGG................................................................................................................................................................................................................................ | 26 | 2 | 14.00 | 14.00 | - | 4.50 | 2.50 | 2.50 | 1.00 | 0.50 | 1.00 | 0.50 | - | 0.50 | - | - | - | - | 0.50 | - | 0.50 | - | - | - |
| .............TGACTGGCTCATTGGCGAGGACTGGGC............................................................................................................................................................................................................................... | 27 | 2 | 13.00 | 13.00 | - | 3.50 | 2.50 | 0.50 | 1.50 | 1.00 | 1.00 | 0.50 | - | 1.50 | - | - | 0.50 | - | - | - | - | - | - | 0.50 |
| .............TGACTGGCTCATTGGCGAGGACTGG................................................................................................................................................................................................................................. | 25 | 2 | 6.00 | 6.00 | - | 1.00 | 1.00 | 1.00 | 0.50 | - | 1.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TGACTGGCTCATTGGCGAGGACTGGGCC.............................................................................................................................................................................................................................. | 28 | 2 | 4.00 | 4.00 | - | - | - | 2.00 | - | 1.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................GTGCTCTGAAACTCCAACCTCCAG.................................................. | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........TCCTGACTGGCTCATTGGCGAGGACT................................................................................................................................................................................................................................... | 26 | 2 | 2.50 | 2.50 | - | 0.50 | 1.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| .............TGACTGGCTCATTGGCGAGGACTGGt................................................................................................................................................................................................................................ | 26 | t | 1.50 | 6.00 | - | 0.50 | - | - | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................TGAGCTGCCCTTGAACCTCACGATTGTGT.... | 29 | 2 | 1.50 | 1.50 | - | - | 0.50 | - | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TGACTGGCTCATTGGCGAGGACTGGGt............................................................................................................................................................................................................................... | 27 | t | 1.50 | 14.00 | - | 0.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GGACAGTCCTGACTGGCTCATTGGCGAGG...................................................................................................................................................................................................................................... | 29 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............TGACTGGCTCATTGGCGAGGACTGGa................................................................................................................................................................................................................................ | 26 | a | 1.00 | 6.00 | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TTGGCGAGGACTGGGCCTTGCTACAGGctgg................................................................................................................................................................................................................ | 31 | ctgg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TGACTGGCTCATTGGCGAGGACTtggc............................................................................................................................................................................................................................... | 27 | tggc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................ATTGGCGAGGACTGGGCCTTGCTACA...................................................................................................................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| ..............GACTGGCTCATTGGCGAGGACTGGGC............................................................................................................................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................TGAGCTGCCCTTGAACCTCACGATTGT...... | 27 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TCCTGACTGGCTCATTGGCGAGGACTtgt................................................................................................................................................................................................................................ | 29 | tgt | 0.50 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ........................TTGGCGAGGACTGGGCCTTG........................................................................................................................................................................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ........AGTCCTGACTGGCTCATTGGCGAGGACTGGG................................................................................................................................................................................................................................ | 31 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GTCCTGACTGGCTCATTGGCGAGGACTGGG................................................................................................................................................................................................................................ | 30 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| ..............................AGGACTGGGCCTTGCTACAG..................................................................................................................................................................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................GCTGCCCTTGAACCTCACGATTGTGTCAC. | 29 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CATTGGCGAGGACTGGGCCTTGCTACAG..................................................................................................................................................................................................................... | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| ............................CGAGGACTGGGCCTTGCTACA...................................................................................................................................................................................................................... | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| .............TGACTGGCTCATTGGCGAGGACTGGGCt.............................................................................................................................................................................................................................. | 28 | t | 0.50 | 13.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GGACAGTCCTGACTGGCTCATTGGCGA........................................................................................................................................................................................................................................ | 27 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..CAGGACAGTCCTGACTGGCTCATTGGC.......................................................................................................................................................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCCAGGACAGTCCTGACTGGCTCATTGGCGAGGACTGGGCCTTGCTACAGGTGAGTGGCTGTGTTATCTTTGAGTTGAGCATTCCTGTGGTTTGTTTGCTCCCCCACATTGGCAATTCATAGTGCTGTGTGAGGTGTTTTTTGACACATGGAAAGACATCAGCAATGCATTCGAAGTGTGTTGACCTGTGTGCTCTGAAACTCCAACCTCCAGGCTGTAAAGCAGTTGCTTGAGCTGCCCTTGAACCTCACGATTGTGTCACC ...................................................................................................................((((.(((((.......((((((((((........))))))))))..((((((((.....))))))))......))))).))))................................................................ .................................................................................................................114................................................................................................213................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................GAGCTGCCCTTGAACCTCACGATTGTGTCA.. | 30 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |