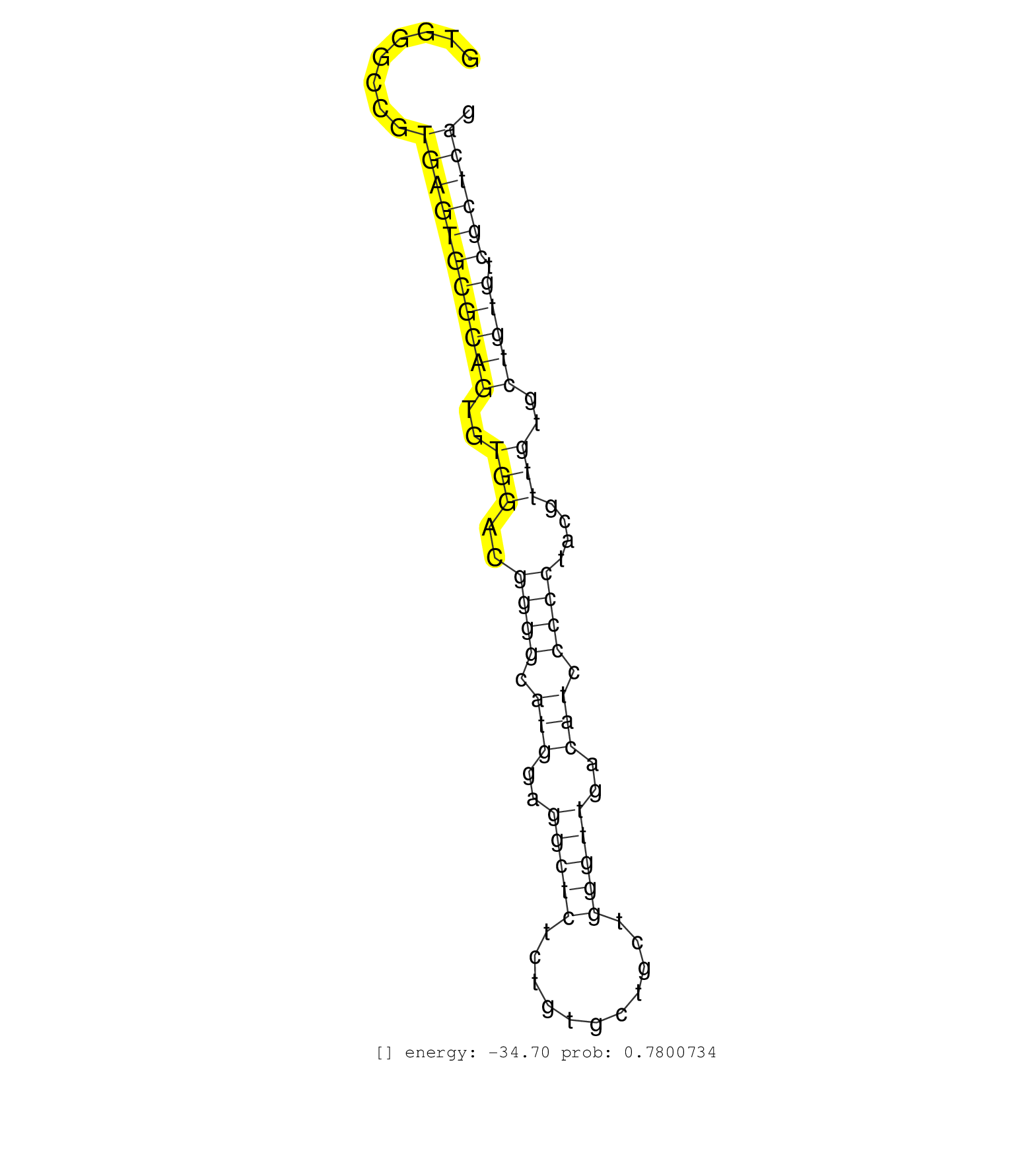

| Gene: Ddx51 | ID: uc008yre.1_intron_0_0_chr5_111082808_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
| GGAGGCTCCTAGGGCGAAGCGGCGGAAAGCGGACAAGGACGTGGACGCAGGTGGGCCGTGAGTGCGCAGTGTGGACGGGGCATGGAGGCTCTCTGTGCTGCTGGGTTGACATCCCCCTACGTTGTGCTGTGTCGCTCAGGGAGAGGTGAGGAGGCTCCGGAGGAGCTCAGTGCCGGCGCAGAGGACCCG ..........................................................(((((((((((..(((..((((.(((..(((((...........)))))..))).))))....)))..))))).))))))................................................... ..................................................51......................................................................................139................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........TAGGGCGAAGCGGCGGAAAGCGGACA.......................................................................................................................................................... | 26 | 1 | 12.00 | 12.00 | 4.00 | 4.00 | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCCGTGAGTGCGCAGTGTGGAC................................................................................................................. | 26 | 1 | 7.00 | 7.00 | - | - | - | - | - | 3.00 | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................TGGGCCGTGAGTGCGCAGTGTGGACGG............................................................................................................... | 27 | 1 | 7.00 | 7.00 | - | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GTGAGTGCGCAGTGTGGACGGGGCATG......................................................................................................... | 27 | 1 | 6.00 | 6.00 | - | - | - | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 |
| ...................................................TGGGCCGTGAGTGCGCAGTGTGGACG................................................................................................................ | 26 | 1 | 6.00 | 6.00 | 2.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................GTGAGTGCGCAGTGTGGACGGGGCAT.......................................................................................................... | 26 | 1 | 5.00 | 5.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGGGCCGTGAGTGCGCAGTGTGGA.................................................................................................................. | 25 | 1 | 5.00 | 5.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................TGAGTGCGCAGTGTGGACGGGGCAT.......................................................................................................... | 25 | 1 | 4.00 | 4.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGAGTGCGCAGTGTGGACGGGGCATG......................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TAGGGCGAAGCGGCGGAAAGCGGAC........................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCCGTGAGTGCGCAGTGTGGACa................................................................................................................ | 27 | a | 3.00 | 7.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCCGTGAGTGCGCAGTGTGGACGt............................................................................................................... | 28 | t | 2.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGGGCCGTGAGTGCGCAGTGTGGAC................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................AAAGCGGACAAGGACGTGGACGCAGGga........................................................................................................................................ | 28 | ga | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........CTAGGGCGAAGCGGCGGAAAGCGGAC........................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GTGAGTGCGCAGTGTGGACGGGGCATt......................................................................................................... | 27 | t | 1.00 | 5.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GTGAGTGCGCAGTGTGGACGGGGt............................................................................................................ | 24 | t | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................AAGGACGTGGACGCAG........................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............GGCGAAGCGGCGGAAAGCGGACAAGG....................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCCGTGAGTGCGCAGTGTGGACGG............................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................AGCGGCGGAAAGCGGACAAGGACGTG.................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........AGGGCGAAGCGGCGGAAAGCGGACA.......................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GTGAGTGCGCAGTGTGGACGGGGCAa.......................................................................................................... | 26 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTGGGCCGTGAGTGCGCAGTGTGG................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTGGGCCGTGAGTGCGCAGTGTGGACag............................................................................................................... | 28 | ag | 1.00 | 7.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCCGTGAGTGCGCAGTGTGGAa................................................................................................................. | 26 | a | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........TAGGGCGAAGCGGCGGAAAGCGGA............................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCCGTGAGTGCGCAGTGTGGt.................................................................................................................. | 25 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GTGAGTGCGCAGTGTGGACGGGG............................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................GTGAGTGCGCAGTGTGGACGGGGC............................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................GGCGGAAAGCGGACAAGGACGTGGACGC............................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................GCGGACAAGGACGTGGACGCAGG.......................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......TCCTAGGGCGAAGCGGCGGAAAGCGGA............................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCCGTGAGTGCGCAGTGTGGACG................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GGAGGAGCTCAGTGCCGG............. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGAGTGCGCAGTGTGGACGGGGCATa......................................................................................................... | 26 | a | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGAGGCTCCTAGGGCGAAGCGGCGGAAAGCGGACAAGGACGTGGACGCAGGTGGGCCGTGAGTGCGCAGTGTGGACGGGGCATGGAGGCTCTCTGTGCTGCTGGGTTGACATCCCCCTACGTTGTGCTGTGTCGCTCAGGGAGAGGTGAGGAGGCTCCGGAGGAGCTCAGTGCCGGCGCAGAGGACCCG ..........................................................(((((((((((..(((..((((.(((..(((((...........)))))..))).))))....)))..))))).))))))................................................... ..................................................51......................................................................................139................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|