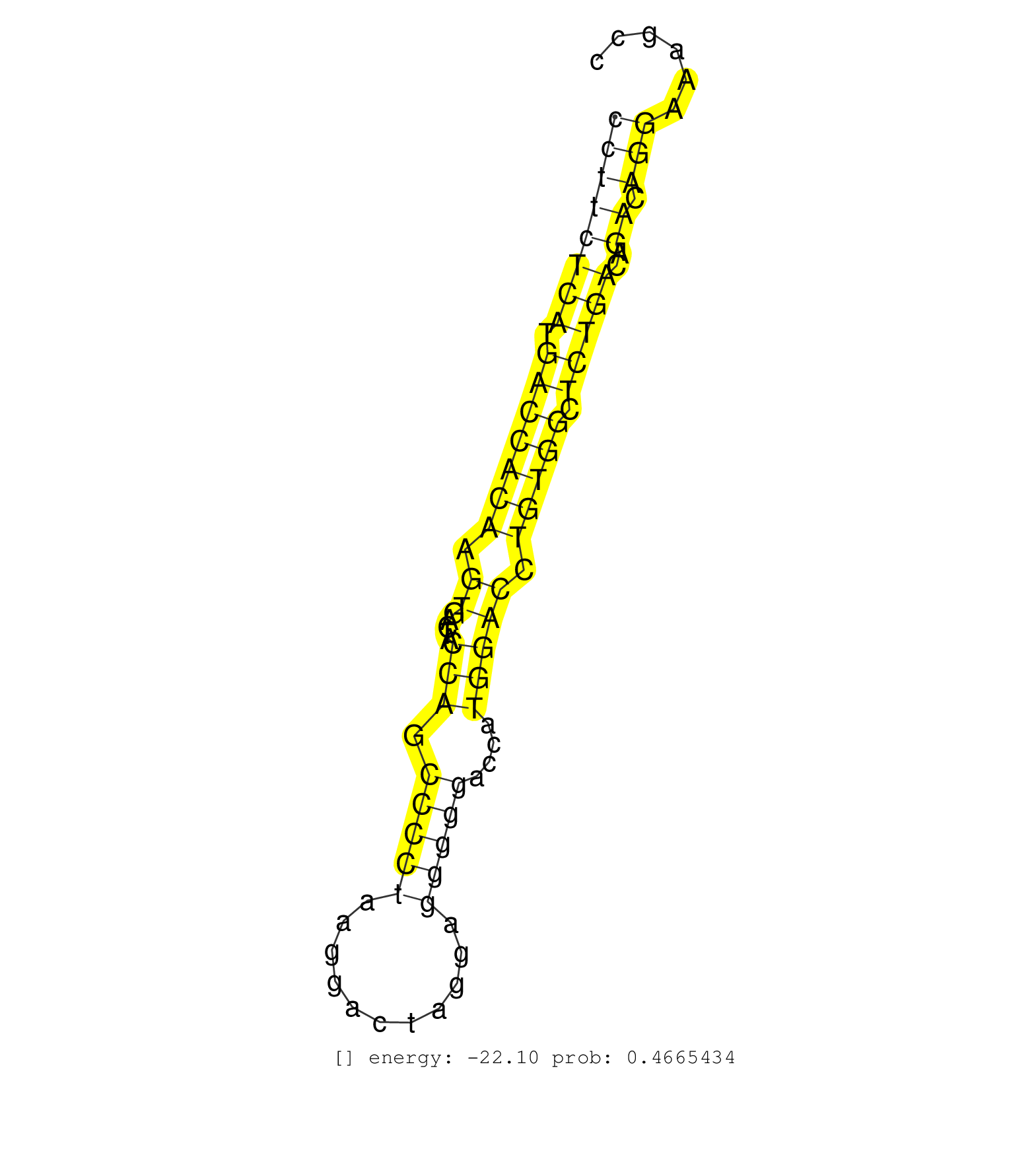

| Gene: Shroom3 | ID: uc008ydt.1_intron_8_0_chr5_93384438_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(5) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(21) TESTES |
| CGGAGGAGCTGCAGGAGGAGGAGGGGCAGGAGGATGTCAATGAGAAAAAGGTAGGGTGGGGACACTTGTTCTGCTTCCCAGGGGAGCCAGGCCCTTCTCATGACCACAAGTGACTACCAGCCCCTAAGGACTAGGAGGGGGACCATGGACCTGTGGCTCTGACAGACAGGAAAGCCACTGTCAGTCTCATTCTGCAAACGATGAATGAAACCTTGGAGAGATTGAGTCATTAGAGGTGTCACAGCTGGGA ............................................................................................((((((((.(((((((.((.....(((.(((((...........)))))....))))).))))).)))))..)).)))................................................................................ ............................................................................................93.................................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................TGGACCTGTGGCTCTGACAGACAGGAA.............................................................................. | 27 | 1 | 46.00 | 46.00 | 14.00 | 12.00 | - | 10.00 | 8.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................................................................................................TGCAAACGATGAATGAAACCTTGGAGA............................... | 27 | 1 | 21.00 | 21.00 | 6.00 | 6.00 | 2.00 | - | - | 1.00 | - | 2.00 | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGGACCTGTGGCTCTGACAGACAGGA............................................................................... | 26 | 1 | 13.00 | 13.00 | 5.00 | 2.00 | - | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................................................................................TGAAACCTTGGAGAGATTGAGTCATT................... | 26 | 1 | 13.00 | 13.00 | 9.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TGCAAACGATGAATGAAACCTTGGAG................................ | 26 | 1 | 12.00 | 12.00 | - | - | 9.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................TCATGACCACAAGTGACTACCAGCCCC.............................................................................................................................. | 27 | 1 | 9.00 | 9.00 | - | 7.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAATGAAACCTTGGAGAGATTGAGTC...................... | 27 | 1 | 9.00 | 9.00 | 2.00 | 1.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................TGAGAAAAAGGTAGGGTGGGGACAC......................................................................................................................................................................................... | 25 | 1 | 9.00 | 9.00 | 3.00 | 4.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TAAGGACTAGGAGGGGGACCATGGA..................................................................................................... | 25 | 1 | 7.00 | 7.00 | - | - | 2.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ........................................TGAGAAAAAGGTAGGGTGGGGACACT........................................................................................................................................................................................ | 26 | 1 | 6.00 | 6.00 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TAAGGACTAGGAGGGGGACC.......................................................................................................... | 20 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TGCAAACGATGAATGAAACCTTGGAGAG.............................. | 28 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGGACCTGTGGCTCTGACAGACAGGAAA............................................................................. | 28 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................AGGATGTCAATGAGAAAAAGGTAGGGTG................................................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................TGTCAATGAGAAAAAGGTAGGGTGGGG............................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGTCAGTCTCATTCTGCAAACGATGAA............................................. | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TCTCATGACCACAAGTGACTACCAGCCCC.............................................................................................................................. | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TGCAAACGATGAATGAAACCTTGGA................................. | 25 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................GAGGATGTCAATGAGAAAAAGGTAGG................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................CGATGAATGAAACCTTGGAGAGATTGA......................... | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TCATGACCACAAGTGACTACCAGCCCCT............................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGGACCTGTGGCTCTGACAGACAGG................................................................................ | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................GAGGATGTCAATGAGAAAAAGGTAGGGT................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................ACTAGGAGGGGGACCATGGACCTGTGGC............................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGAGAAAAAGGTAGGGTGGGGAC........................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGAGAAAAAGGTAGGGTGGGGACAa......................................................................................................................................................................................... | 25 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGAGAAAAAGGTAGGGTGGGGACACTT....................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CATGACCACAAGTGACTACCAGCCCCT............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAATGAAACCTTGGAGAGATTGAG........................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................AGGAGGGGGACCATGGACCTGTGGC............................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................TGTCAATGAGAAAAAGGTAGG................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................TGTCAATGAGAAAAAGGTAGGGTGGGt............................................................................................................................................................................................. | 27 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................GTCATTAGAGGTGTCACAGCTGGa. | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGAGTCATTAGAGGTGTCACAGCTGG.. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GACCTGTGGCTCTGACAGACAGGAAA............................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TGACTACCAGCCCCTAAGGACTAGGAGG................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................TGTCAATGAGAAAAAGGTAGGGTGtgg............................................................................................................................................................................................. | 27 | tgg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGGAGAGATTGAGTCATTAGAGGTGT........... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGGACCTGTGGCTCTGACAG..................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TAAGGACTAGGAGGGGGACCATGG...................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGGACCTGTGGCTCTGACAGACAGGAt.............................................................................. | 27 | t | 1.00 | 13.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGGAGAGATTGAGTCATTAGAGGTG............ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TGACCACAAGTGACTACCAGCCCCTAA........................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TAAGGACTAGGAGGGGGACCATGGg..................................................................................................... | 25 | g | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TCTGCAAACGATGAATGAAACCTTGGA................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAAACCTTGGAGAGATTGAGTCA..................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAAACCTTGGAGAGATTGAGTCAT.................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TTCTCATGACCACAAGTGACTACCAGacc............................................................................................................................... | 29 | acc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................CATTCTGCAAACGATGAATGAAACCTTG................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................TCATGACCACAAGTGACTACCAGCCC............................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................AGTGACTACCAGCCCCTAAGGACTAGG................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TGCAAACGATGAATGAAACCTTGGAGAGA............................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TGCAAACGATGAATGAAACCTTGGAGt............................... | 27 | t | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................TTCTCATGACCACAAGTGACTACCAGCCCC.............................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TCTGCAAACGATGAATGAAACCTTGGAGA............................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| CGGAGGAGCTGCAGGAGGAGGAGGGGCAGGAGGATGTCAATGAGAAAAAGGTAGGGTGGGGACACTTGTTCTGCTTCCCAGGGGAGCCAGGCCCTTCTCATGACCACAAGTGACTACCAGCCCCTAAGGACTAGGAGGGGGACCATGGACCTGTGGCTCTGACAGACAGGAAAGCCACTGTCAGTCTCATTCTGCAAACGATGAATGAAACCTTGGAGAGATTGAGTCATTAGAGGTGTCACAGCTGGGA ............................................................................................((((((((.(((((((.((.....(((.(((((...........)))))....))))).))))).)))))..)).)))................................................................................ ............................................................................................93.................................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................CTTGTTCTGCTTCCCAG......................................................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - |