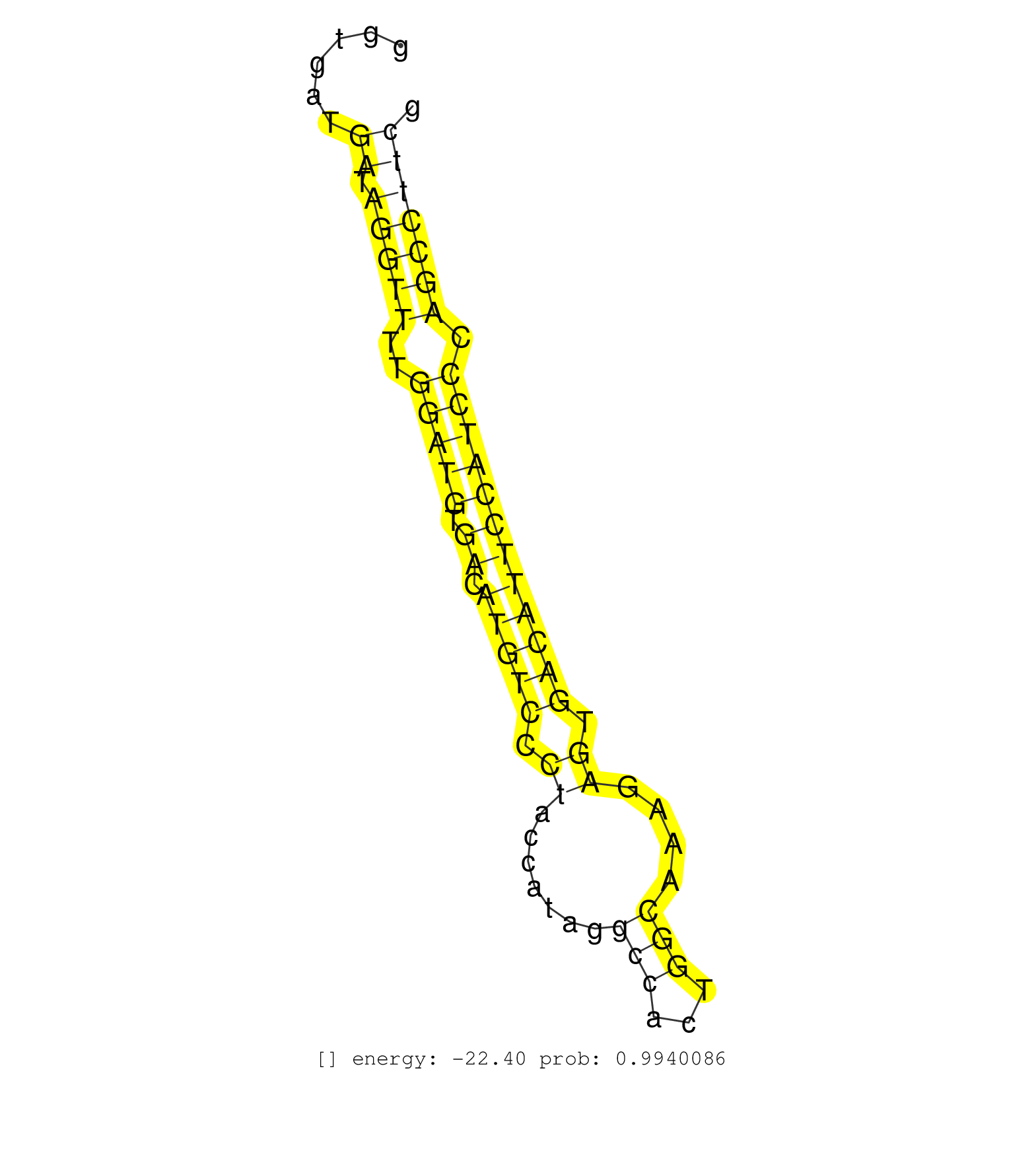

| Gene: Shroom3 | ID: uc008ydt.1_intron_7_0_chr5_93382057_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| AGGAAAGCCTTAGACATCACCGCCAGGCGTGCGGGATGTGAAGCCAAGGCGTAAGTCCCCTCTAGTCCACCCCAAAGAGCCTGTGACTCAACTCCTGTGGGGTGATGATAGGTTTTGGATGTGACATGTCCCTACCATAGGCCACTGGCAAAGAGTGACATTCCATCCCAGCCTTCGGGCCGGAGGTGGGGCAGATGCCCAACAGCAAGAATGAGAGCAGTAGGAACCACTGTTATTTGTAGCCAGAGGT ..........................................................................................................((.(((((..(((((.((.(((((.((.......(((...)))....)).)))))))))))).))))))).......................................................................... ....................................................................................................101.........................................................................177....................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................TGATAGGTTTTGGATGTGACATGTCCC...................................................................................................................... | 27 | 1 | 6.00 | 6.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................TGTGGGGTGATGATAGGTTTTGGATGT................................................................................................................................ | 27 | 1 | 4.00 | 4.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................CGGGATGTGAAGCCAAGGCGagc.................................................................................................................................................................................................... | 23 | agc | 3.00 | 0.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGATAGGTTTTGGATGTGACATGTCCCT..................................................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................TGATAGGTTTTGGATGTGACATGTCC....................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................CAGCAAGAATGAGAGCAGTAGGAACC...................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGGCAAAGAGTGACATTCCATCCCAGCC............................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................GGAACCACTGTTATTTGTAGCCAGAGG. | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CTCTAGTCCACCCCAgtga............................................................................................................................................................................ | 19 | gtga | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 |
| ......................................................................................................................................................................................................CCAACAGCAAGAATGAGAGCAGTAGGA......................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................CAACAGCAAGAATGAGAGCAGTAGG.......................... | 25 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TGAGAGCAGTAGGAACCACTGTTATT............. | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CAAGAATGAGAGCAGTAGGAACCACTG.................. | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGATAGGTTTTGGATGTGACATGTCCCTA.................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................TACCATAGGCCACTGGCAAAGAGTG............................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGCCCAACAGCAAGAATGAGAGCAGTAGG.......................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGTGACTCAACTCCTGTGGGGTGATG............................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AGGCGTGCGGGATGTGAAGCCAAGGCG....................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TAGGAACCACTGTTATTTGTAGCCAGA... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................................................................................................................CCCAACAGCAAGAATGAGAGCAGTAG........................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................AACAGCAAGAATGAGAGCAGTAGGA......................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TAGACATCACCGCCAGGCGTGCGGGAcg.................................................................................................................................................................................................................... | 28 | cg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TAGACATCACCGCCAGGCGTGCGGG....................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................AGCAAGAATGAGAGCAGTAGGAACCAC.................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................AGCCTGTGACTCAACTCCTGTGGGGTGt................................................................................................................................................. | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................GGTGATGATAGGTTTTGGATGTGACATG.......................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................CGCCAGGCGTGCGGGATGTGt................................................................................................................................................................................................................. | 21 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................................................CAGCAAGAATGAGAGCAGTAGGAACCA..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGATGATAGGTTTTGGATGTGACAT........................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGTGACTCAACTCCTGTGGGGTGATGA.............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TACCATAGGCCACTGGCAAAGAGTGA............................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TAGGTTTTGGATGTGACATGTCCCTA.................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGCCCAACAGCAAGAATGAGAGCAGT............................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................TGGGGCAGATGCCCAACAGCAAGAATGAGAG................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGTGGGGTGATGATAGGTTTTGGATGTG............................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................ACAGCAAGAATGAGAGCAGTAGGAAC....................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CAAGAATGAGAGCAGTAGGAACCAC.................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGCCCAACAGCAAGAATGAGAGC................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................GCAGTAGGAACCACTG.................. | 16 | 6 | 0.17 | 0.17 | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGAAAGCCTTAGACATCACCGCCAGGCGTGCGGGATGTGAAGCCAAGGCGTAAGTCCCCTCTAGTCCACCCCAAAGAGCCTGTGACTCAACTCCTGTGGGGTGATGATAGGTTTTGGATGTGACATGTCCCTACCATAGGCCACTGGCAAAGAGTGACATTCCATCCCAGCCTTCGGGCCGGAGGTGGGGCAGATGCCCAACAGCAAGAATGAGAGCAGTAGGAACCACTGTTATTTGTAGCCAGAGGT ..........................................................................................................((.(((((..(((((.((.(((((.((.......(((...)))....)).)))))))))))).))))))).......................................................................... ....................................................................................................101.........................................................................177....................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................CCTTCGGGCCGGAacca.................................................................. | 17 | acca | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ......................GCGTGCGGGATgccc..................................................................................................................................................................................................................... | 15 | gccc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................CCCCTCTAGTCCACC................................................................................................................................................................................... | 15 | 11 | 0.36 | 0.36 | - | - | - | - | - | - | 0.36 | - | - | - | - | - | - | - | - | - | - | - | - |