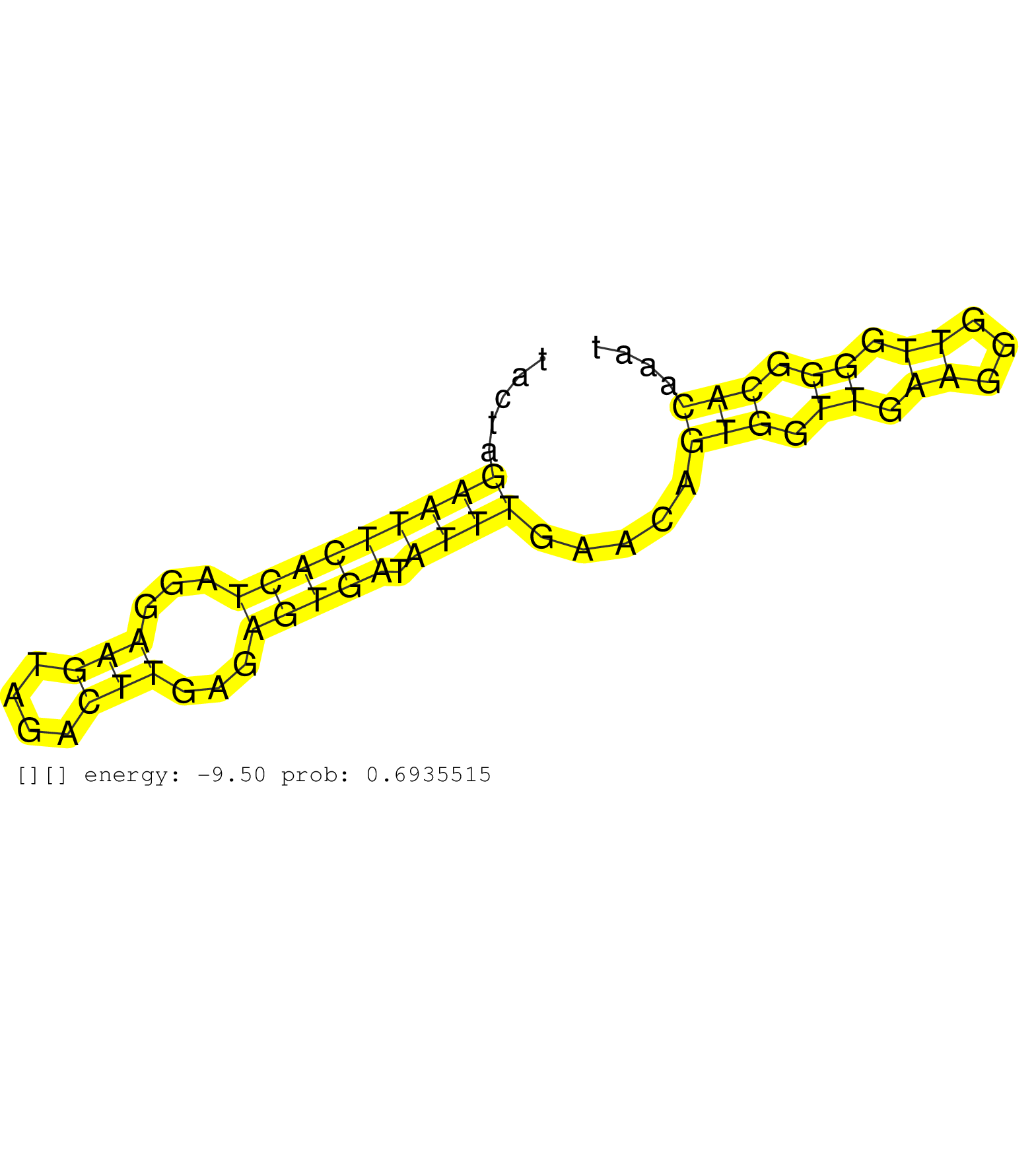

| Gene: Clock | ID: uc008xuq.1_intron_14_0_chr5_76677488_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| GTATAATATATGTATCTGAGAGTGTAACTTCGTTACTTGAACATTTACCAGTAAGTATGAAAACCCTATAATCTCTATTATTTAATGTCTATATCACATACTAGAATTCACTAGGAAGTAGACTTGAGAGTGATATTTGAACAGTGGTTGAAGGGTTGGGGCACAAATTCCAGTTGTGTCTTTATTAGTGTTGCTGTTTCTCAAACTAGTCCTAAGTTCTTCATTCTTAGCACCTAGTAATTTAACCTGC .......................................................................................................(((((((((...(((....)))...))))).)))).....(((.((.((...)).)).)))...................................................................................... ..................................................................................................99...................................................................168................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................GAATTCACTAGGAAGTAGACTTGAGAGTGATATTTGAACAGTGGTTGAAGGG............................................................................................... | 52 | 1 | 35.00 | 35.00 | 35.00 | - | - | - | - | - | - | - | - | - | - |
| ...TAATATATGTATCTGttt..................................................................................................................................................................................................................................... | 18 | ttt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........TGTATCTGAGAGTGTAACTTCGTTAt...................................................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .TATAATATATGTATCTGAGAGTGTAAC.............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....AATATATGTATCTGAGAGTGTAACT............................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACAGTGGTTGAAGGGTTGGGGCAC...................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........ATGTATCTGAGAGTGTAACTTCGTTACT..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..ATAATATATGTATCTGAGAG.................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................................GGTTGGGGCACAAAaaa................................................................................ | 17 | aaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................AACTTCGTTACTTGAACATTTACCAtctg.................................................................................................................................................................................................... | 29 | tctg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| GTATAATATATGTATCTGAGAGTGTAACTTCGTTACTTGAACATTTACCAGTAAGTATGAAAACCCTATAATCTCTATTATTTAATGTCTATATCACATACTAGAATTCACTAGGAAGTAGACTTGAGAGTGATATTTGAACAGTGGTTGAAGGGTTGGGGCACAAATTCCAGTTGTGTCTTTATTAGTGTTGCTGTTTCTCAAACTAGTCCTAAGTTCTTCATTCTTAGCACCTAGTAATTTAACCTGC .......................................................................................................(((((((((...(((....)))...))))).)))).....(((.((.((...)).)).)))...................................................................................... ..................................................................................................99...................................................................168................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................................TCTTAGCACCTAGTAA.......... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | 0.25 |