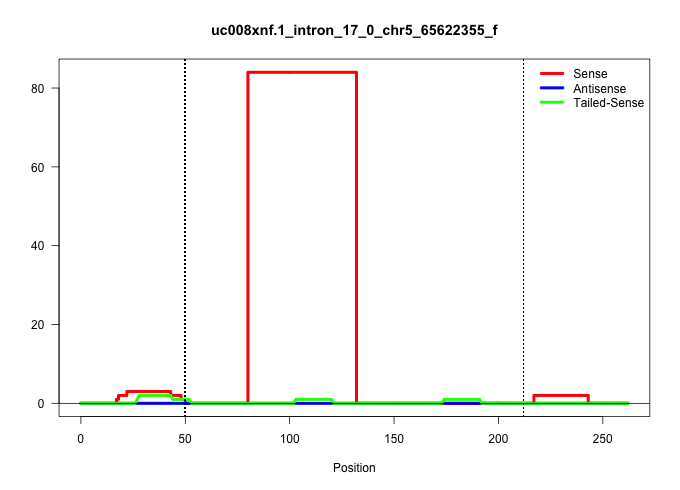
| Gene: Wdr19 | ID: uc008xnf.1_intron_17_0_chr5_65622355_f | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(9) TESTES |
| CCCGGACAATGGGGGATGTTGGCACAGTGATGTCGTTGGAACAAATAAAGGTATGCAGTGTTTTAGGACTCACTAGATCACAGCATAGGTGCTGCTTATTCGGCATTTTACGTTTTAGTCTCAATTCCAACAACTGATACTTCCTACCGATCAAAGCCTTTGAACAATAAAATCCTTATATCTCTTCTCATCGCAACTGGTTTTCTTTGTAGGGAATCGAGGACTACAATCTTTTGGCAGGACATCTCGCCATGTTTACTAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................CAGCATAGGTGCTGCTTATTCGGCATTTTACGTTTTAGTCTCAATTCCAACA.................................................................................................................................. | 52 | 1 | 84.00 | 84.00 | 84.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................CGAGGACTACAATCTTTTGGCAGGAC................... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - |
| ......................CACAGTGATGTCGTTGGAACAAATAAAG.................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................CTTATATCTCTTCTgaac...................................................................... | 18 | gaac | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - |
| ...........................TGATGTCGTTGGAACAAATAAAGGga................................................................................................................................................................................................................. | 26 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................CATTTTACGTTTTAaagt............................................................................................................................................. | 18 | aagt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - |
| ............................GATGTCGTTGGAtaaa.......................................................................................................................................................................................................................... | 16 | taaa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - |
| .................GTTGGCACAGTGATGTCGTTGGAACA........................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ..................TTGGCACAGTGATGTCGTTGGAACAAATAA...................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| CCCGGACAATGGGGGATGTTGGCACAGTGATGTCGTTGGAACAAATAAAGGTATGCAGTGTTTTAGGACTCACTAGATCACAGCATAGGTGCTGCTTATTCGGCATTTTACGTTTTAGTCTCAATTCCAACAACTGATACTTCCTACCGATCAAAGCCTTTGAACAATAAAATCCTTATATCTCTTCTCATCGCAACTGGTTTTCTTTGTAGGGAATCGAGGACTACAATCTTTTGGCAGGACATCTCGCCATGTTTACTAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................CGATCAAAGCCTTtaga...................................................................................................... | 17 | taga | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - |