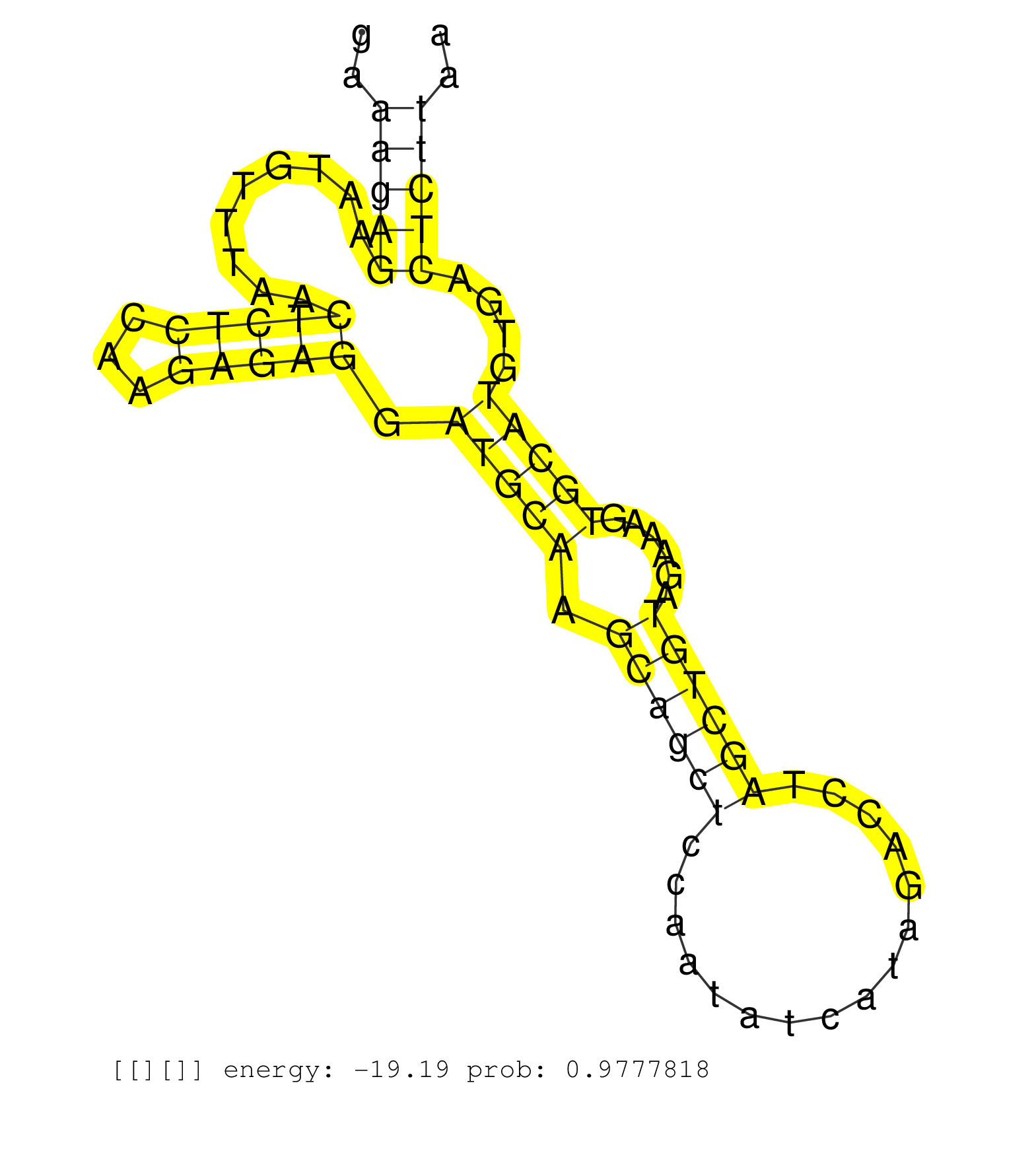

| Gene: Hdh | ID: uc008xda.1_intron_2_0_chr5_35136814_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(15) TESTES |
| CCCTCTGGATTTTAAAAATCTTTGAAGTAATTACCATCTATTTCACACTGTTAATTTTGACTTTCTCTGATTAAATTAACAGAGAAAGAGAATGTTTAACTCTCCAAGAGAGGATGCAAGCAGCTCCAATATCATAGACCTAGCTGTAGAAAGTGCATGTGACTCTTAAATGTAACACATCACTTGTTAACTCCACTTAGAATGGTGCTCCTCGAAGTTTGCGTGCTGCCCTGTGGAGGTTTGCTGAGCT .....................................................................................(((((.........(((((...))))).(((((.((((((................))))))......)))))....)))))................................................................................... ...................................................................................84...................................................................................169............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................GACCTAGCTGTAGAAAGTGCATGTGACTC..................................................................................... | 29 | 1 | 5.00 | 5.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| .........................................................................................................................................ACCTAGCTGTAGAAAGTGCATGTGACT...................................................................................... | 27 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................................GACCTAGCTGTAGAAAGTGCATGTGACT...................................................................................... | 28 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGCGTGCTGCCCTGTGGAGGTTTGCTGAGC. | 30 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ACCTAGCTGTAGAAAGTGCATGTGACTC..................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CGAAGTTTGCGTGCTGCCCTGTGGAG............ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................AGAATGTTTAACTCTCCAAGAGAGGATGCAAGC................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TCATAGACCTAGCTGTAGAAAGTGCA............................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGCTCCTCGAAGTTTGCGTGCTGCCCT.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GACCTAGCTGTAGAAAGTGCATGTGACTCT.................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGGTGCTCCTCGAAGTTTGCGTGCTGC..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................ATCATAGACCTAGCTGTAGAAAGTGCAT............................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGGTGCTCCTCGAAGTTTGCGTGCTG...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ACCTAGCTGTAGAAAGTGCATGTGACTCTTAAAT............................................................................... | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................AATGTTTAACTCTCCAAGAGAGGATGC..................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................TCATAGACCTAGCTGTAGAAAGTGCATG........................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TAGACCTAGCTGTAGAAAGTGCATGTGA........................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TCTCCAAGAGAGGATGCAAGCAGCTCCcaa........................................................................................................................ | 30 | caa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................TCATAGACCTAGCTGTAGAAAGTGCAT............................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................ACCTAGCTGTAGAAAGTGCATGTGACTCT.................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| CCCTCTGGATTTTAAAAATCTTTGAAGTAATTACCATCTATTTCACACTGTTAATTTTGACTTTCTCTGATTAAATTAACAGAGAAAGAGAATGTTTAACTCTCCAAGAGAGGATGCAAGCAGCTCCAATATCATAGACCTAGCTGTAGAAAGTGCATGTGACTCTTAAATGTAACACATCACTTGTTAACTCCACTTAGAATGGTGCTCCTCGAAGTTTGCGTGCTGCCCTGTGGAGGTTTGCTGAGCT .....................................................................................(((((.........(((((...))))).(((((.((((((................))))))......)))))....)))))................................................................................... ...................................................................................84...................................................................................169............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|