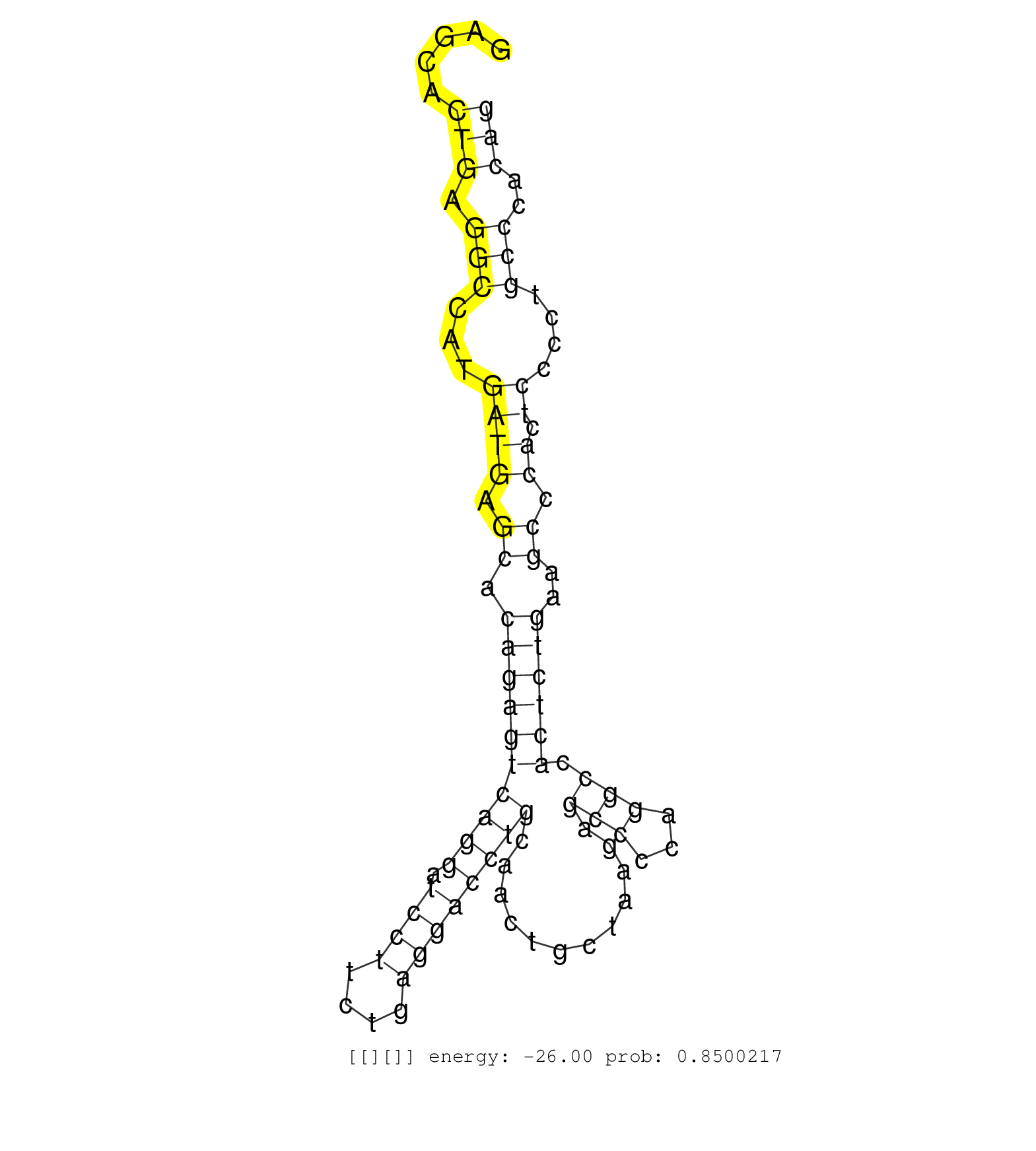

| Gene: mKIAA1643 | ID: uc008xcb.1_intron_4_0_chr5_34577137_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(23) TESTES |
| CACAACCCTGAGATGCTCTTGGCTCCCTCCCAGCAGCCACCCTGGACCAGGAGGCAATGAGAGTCACATTACTGGGTCCCTCTGTATTGAATCCCACACTGAGCACTGAGGCCATGATGAGCACAGAGTCAGGATCCTTCTGAGGACCTGCAACTGCTAAGAGCCCCAGGCCACTCTGAAGCCCACTCCCCTGCCCACAGTGCCTGGCAGCTGGCTCCATCATCATGAATCGGGAGCTGGAGAGCATGGC .........................................................................................................(((.(((...((((.((.((((((((((.((((....))))))))............(((...))).))))))..)).)).))....)))..))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................CACACTGAGCACTGAGGCCATGATGAG................................................................................................................................. | 27 | 1 | 14.00 | 14.00 | 9.00 | - | - | - | - | - | 1.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TCATGAATCGGGAGCTGGAGAGCATG.. | 26 | 1 | 12.00 | 12.00 | 1.00 | 3.00 | 1.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TCATGAATCGGGAGCTGGAGAGCAT... | 25 | 1 | 9.00 | 9.00 | - | 1.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................CACACTGAGCACTGAGGCCATGATGAGC................................................................................................................................ | 28 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................CCATCATCATGAATCGGGAGCTGGAG........ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................CACACTGAGCACTGAGGCCATGATGgg................................................................................................................................. | 27 | gg | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................................................................................................TCATCATGAATCGGGAGCTGGAGAGC..... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TGAATCGGGAGCTGGAGAGC..... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................CCACACTGAGCACTGAGGCCATGATGAG................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................CCATCATCATGAATCGGGAGCTGGAGAGC..... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................CACACTGAGCACTGAGGCCATGATcag................................................................................................................................. | 27 | cag | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................ACACTGAGCACTGAGGCCATGATGAG................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TGAATCGGGAGCTGGAGAGCATGGt | 25 | t | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TGAATCGGGAGCTGGAGAGCATGGC | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TGAATCGGGAGCTGGAGAGCATG.. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CCCACACTGAGCACTGAGGCCATGATGAG................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................ATGAATCGGGAGCTGGAGAGCATGGC | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TGAATCGGGAGCTGGAGAGCATGG. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGAGGACCTGCAACTGCTAAGAGCCCC................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................CACACTGAGCACTGAGGCCATGATGAGCAC.............................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................................CATCATGAATCGGGAGCTGGAGAGC..... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................CACACTGAGCACTGAGGCCATGATGg.................................................................................................................................. | 26 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................................................................TGAATCGGGAGCTGGAGAGCATGGCt | 26 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGGCAGCTGGCTCCATCATCATGAATC................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TCCCACACTGAGCACTGAGGCCATGATG................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................ATCGGGAGCTGGAGAGCAT... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CACAACCCTGAGATGCTCTTGGCTCCCTCCCAGCAGCCACCCTGGACCAGGAGGCAATGAGAGTCACATTACTGGGTCCCTCTGTATTGAATCCCACACTGAGCACTGAGGCCATGATGAGCACAGAGTCAGGATCCTTCTGAGGACCTGCAACTGCTAAGAGCCCCAGGCCACTCTGAAGCCCACTCCCCTGCCCACAGTGCCTGGCAGCTGGCTCCATCATCATGAATCGGGAGCTGGAGAGCATGGC .........................................................................................................(((.(((...((((.((.((((((((((.((((....))))))))............(((...))).))))))..)).)).))....)))..))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................CTCCATCATCATGAAggt..................... | 18 | ggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |