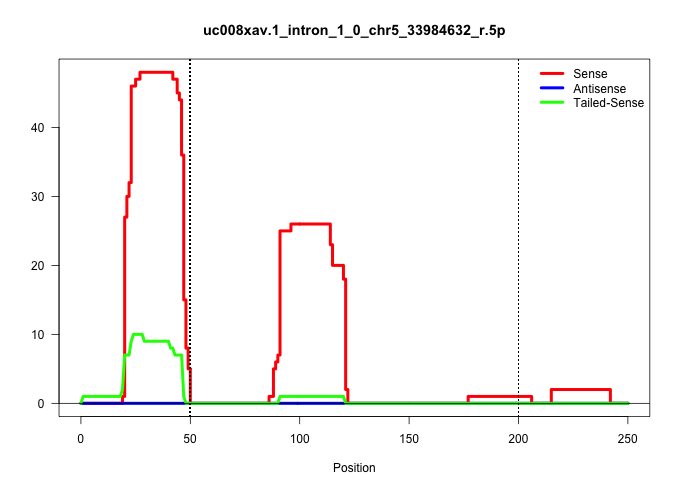
| Gene: Slbp | ID: uc008xav.1_intron_1_0_chr5_33984632_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(18) TESTES |
| TTGCATTTTTGGGATCCTCCTGCTGAAGAAGGATGTGATTTGCAAGAAATGTATGTCTTTCTTTATCTCTTGTCACAGTGACAGGTTATCCTGAACATTGCTGAGAACAGGCATTAGCCTTTAGGCCTGATCTATTACACAGAGTTGTGGATGCCAGGGAGGAGTTTACCTCCTGTTCTGTATTTAGTCTTTGAGAGTTAGAAAACAGCTCAGTCTAGGACAATTGGCTGTGTGTGCACTTCAGAGGACA ...................................................................................................((((....(((((............))))).((((.....(((((...(((((((..((((((......)))))).....)))))))..))))).....))))...))))......................................... ......................................................................................87.........................................................................................................................210...................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................TGCTGAAGAAGGATGTGATTTGCAAGA........................................................................................................................................................................................................... | 27 | 1 | 19.00 | 19.00 | 5.00 | 4.00 | 4.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................TGAACATTGCTGAGAACAGGCATTAGCCTT................................................................................................................................. | 30 | 1 | 14.00 | 14.00 | 3.00 | 4.00 | - | 1.00 | - | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................TGCTGAAGAAGGATGTGATTTGCAAG............................................................................................................................................................................................................ | 26 | 1 | 6.00 | 6.00 | 1.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGAAGAAGGATGTGATTTGCAAGAAAT........................................................................................................................................................................................................ | 27 | 1 | 5.00 | 5.00 | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................TGCTGAAGAAGGATGTGATTTGCAAGt........................................................................................................................................................................................................... | 27 | t | 4.00 | 6.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGAAGAAGGATGTGATTTGCAAGAA.......................................................................................................................................................................................................... | 25 | 1 | 4.00 | 4.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................TCCTGAACATTGCTGAGAACAGGCAT........................................................................................................................................ | 26 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CTGAAGAAGGATGTGATTTGCAAGA........................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................TGAACATTGCTGAGAACAGGCATTAGCCTTT................................................................................................................................ | 31 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| .............................AGGATGTGATTTGCAAGA........................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TAGGACAATTGGCTGTGTGTGCACTTC........ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................TGAACATTGCTGAGAACAGGCATTAGCCT.................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGAAGAAGGATGTGATTTGCA.............................................................................................................................................................................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGAAGAAGGATGTGATTTGCAAGAAA......................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................GCTGAAGAAGGATGTGATTTGCAAG............................................................................................................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CTGTATTTAGTCTTTGAGAGTTAGAAAAC............................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................AAGAAGGATGTGATTTGCAAGAA.......................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TATCCTGAACATTGCTGAGAACAGGCATT....................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGAAGAAGGATGTGATTTGt............................................................................................................................................................................................................... | 20 | t | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................GAAGGATGTGATTTGCAAGAA.......................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................ATTGCTGAGAACAGGCATTAGCCTT................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TGCTGAAGAAGGATGTGATTTGCAAGAt.......................................................................................................................................................................................................... | 28 | t | 1.00 | 19.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CCTGAACATTGCTGAGAACAGGCATTAGCCTT................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................CTGAACATTGCTGAGAACAGGCATT....................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................GCTGAAGAAGGATGTGATTTGCAAGAA.......................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TGCTGAAGAAGGATGTGATTTGCAAGc........................................................................................................................................................................................................... | 27 | c | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................TCCTGAACATTGCTGAGAACAGGCATT....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGAAGAAGGATGTGATTTGCAAGt........................................................................................................................................................................................................... | 24 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................CTGCTGAAGAAGGATGTGATTTGCAAGt........................................................................................................................................................................................................... | 28 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GAAGAAGGATGTGATTa................................................................................................................................................................................................................. | 17 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CTGCTGAAGAAGGATGTGATTTGCAA............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TGCATTTTTGGGATCCTCCTGCTGAAGt............................................................................................................................................................................................................................. | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................TGCTGAAGAAGGATGTGATTTGCAAGAAA......................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGAACATTGCTGAGAACAGGCATTAGactt................................................................................................................................. | 30 | actt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGAAGAAGGATGTGATTTG................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................CTGAACATTGCTGAGA................................................................................................................................................ | 16 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - |
| TTGCATTTTTGGGATCCTCCTGCTGAAGAAGGATGTGATTTGCAAGAAATGTATGTCTTTCTTTATCTCTTGTCACAGTGACAGGTTATCCTGAACATTGCTGAGAACAGGCATTAGCCTTTAGGCCTGATCTATTACACAGAGTTGTGGATGCCAGGGAGGAGTTTACCTCCTGTTCTGTATTTAGTCTTTGAGAGTTAGAAAACAGCTCAGTCTAGGACAATTGGCTGTGTGTGCACTTCAGAGGACA ...................................................................................................((((....(((((............))))).((((.....(((((...(((((((..((((((......)))))).....)))))))..))))).....))))...))))......................................... ......................................................................................87.........................................................................................................................210...................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|