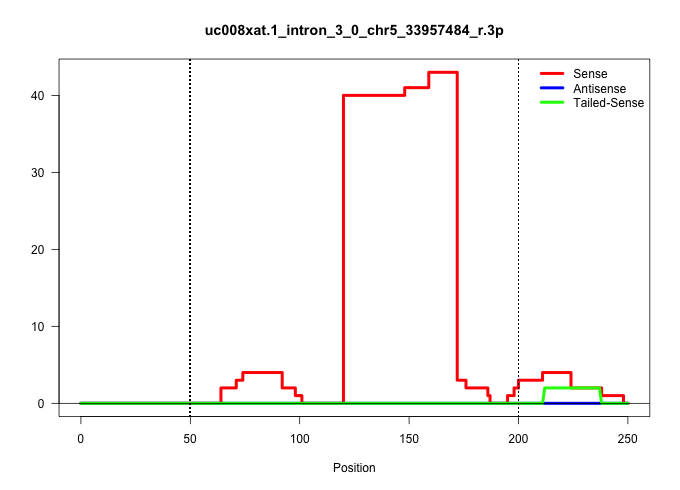
| Gene: 2410018C17Rik | ID: uc008xat.1_intron_3_0_chr5_33957484_r.3p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(11) TESTES |
| TTCAACCCCCAAAGGTGAGAACCACTATTCTAGAAGATATTGCTAGAGGCTTCAGGAAGGAACATACTTGGTTATCTTAGAATTAGGAAGGCATTTGTCAAAGAGGGATATTAGGAACATTTTTTAAAAACAGGATGTTGTATATATATAACAGGAATATGGACTGAAGTGTATGGTCATGTGGCAATTTCTCTTTACAGAGACAAAGCCCTGAAGAGTCTGGAAATGGGTCCGCATCTTTAAGTCAGAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................TTTTTAAAAACAGGATGTTGTATATATATAACAGGAATATGGACTGAAGTGT.............................................................................. | 52 | 1 | 40.00 | 40.00 | 40.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................TACTTGGTTATCTTAGAATTAGGAAGGC.............................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................GAAGAGTCTGGAAATGGGTCCGCATa............ | 26 | a | 2.00 | 0.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TGGACTGAAGTGTATGGTCATGTGGCA................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................AATGGGTCCGCATCTTTAAGTCAG.. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................................................TACAGAGACAAAGCCCTGAAGAGTCTGGA.......................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGAGACAAAGCCCTGAAGAGTCTGGA.......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................................AGACAAAGCCCTGAAGAGTCTGGA.......................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................................................................TGAAGAGTCTGGAAATGGGTCCGCATC............ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................TGGACTGAAGTGTATGGTCATGTGGCAA............................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................TCTTAGAATTAGGAAGGCATTTGTCAA..................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TAACAGGAATATGGACTGAAGTGTATGG.......................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................TTATCTTAGAATTAGGAAGGCATTTGT........................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| TTCAACCCCCAAAGGTGAGAACCACTATTCTAGAAGATATTGCTAGAGGCTTCAGGAAGGAACATACTTGGTTATCTTAGAATTAGGAAGGCATTTGTCAAAGAGGGATATTAGGAACATTTTTTAAAAACAGGATGTTGTATATATATAACAGGAATATGGACTGAAGTGTATGGTCATGTGGCAATTTCTCTTTACAGAGACAAAGCCCTGAAGAGTCTGGAAATGGGTCCGCATCTTTAAGTCAGAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|