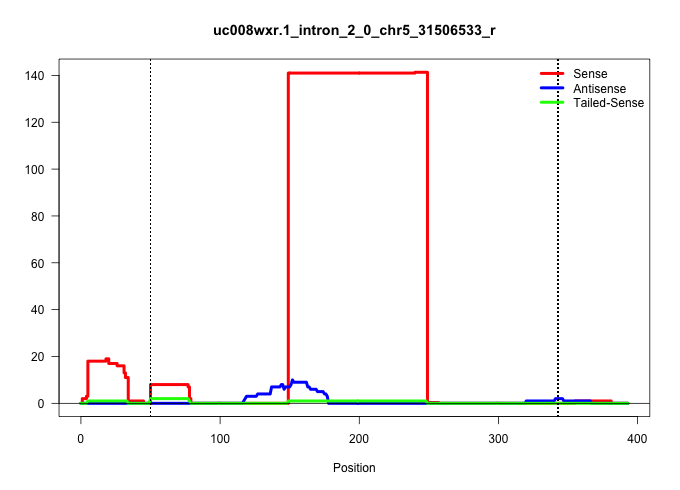
| Gene: Ppm1g | ID: uc008wxr.1_intron_2_0_chr5_31506533_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(1) TDRD1.ip |
(22) TESTES |
| CACCATGGATGGACGAGTCAATGGAGGCCTCAACCTCTCCAGGGCCATTGGTAAGGACTAAGAAACTAGGCAATCTTGGGGTTTTGTCCTGTGTCCCAAACTACCAATAGCAAAGTTTAAGCCCTGTGCTCTTCAGAATAAGCTCATGCCTTCTTTGTTCTTAAATACCTCTGTAAAATGGTAAAGTTATACTTTTCCCTGTGTTCTGGATGTTAAAAGAGGCAAATGAGCTCTTTGTTCTGAGAGCAGATCTGAAACTAGAAAAGATGAAACTTAAAGATTTGACCCACCTCTAATACAAGTCTGGGAATTAGTCAAACTACTCACCGTCCTTACTTTTCAGGAGACCACTTCTACAAGAGAAACAAAAACTTGCCACCCCAGGAACAGATG .....................................................(((((.((((..(((((....)))))..))))))))).......(((((...........)))))........(((((.(((((((((((((((((((..((((.(((..(((((...........((.((((.....)))).))..)))))..)))...))))..)))))..))))))...)))))))))))))................................................................................................................................................. ..................................................51........................................................................................................................................................................................................253.......................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................CTTCTTTGTTCTTAAATACCTCTGTAAAATGGTAAAGTTATACTTTTCCCTG................................................................................................................................................................................................ | 52 | 1 | 156.00 | 156.00 | 156.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGGATGGACGAGTCAATGGAGGCCTCAAC....................................................................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 10.00 | 10.00 | - | 7.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACTAAGAAACTAGGCAATCTTG........................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 5.00 | 5.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAAGGACTAAGAAACTAGGCAATCTTGG.......................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .ACCATGGATGGACGAGTCA..................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGGATGGACGAGTCAATGGAGGCCTCA......................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....TGGATGGACGAGTCAATGGAGGCCTC.......................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACTAAGAAACTAGGCAATCTTt........................................................................................................................................................................................................................................................................................................................... | 28 | t | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACTAAGAAACTAGGCAATCTT............................................................................................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................TTCTACAAGAGAAACAAAAA...................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....TGGATGGACGAGTCAATGGAGGCCTCAAa....................................................................................................................................................................................................................................................................................................................................................................... | 29 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................AGATGAAACTTAAAGgaa............................................................................................................... | 18 | gaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTAAGGACTAAGAAACTAGGCAATCTTGt.......................................................................................................................................................................................................................................................................................................................... | 29 | t | 1.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................CAATGGAGGCCTCAACCTCTCCAGGGC............................................................................................................................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....ATGGATGGACGAGTCAATGGAGGCCTC.......................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................ACAAGAGAAACAAAAACTTGCCACCC............ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................................................GTTCTGGATGTTAAAAGAGGCAAATGAGC.................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................................................................................................................................................TGAGAGCAGATCTGAAA........................................................................................................................................ | 17 | 3 | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CACCATGGATGGACGAGTCAATGGAGGCCTCAACCTCTCCAGGGCCATTGGTAAGGACTAAGAAACTAGGCAATCTTGGGGTTTTGTCCTGTGTCCCAAACTACCAATAGCAAAGTTTAAGCCCTGTGCTCTTCAGAATAAGCTCATGCCTTCTTTGTTCTTAAATACCTCTGTAAAATGGTAAAGTTATACTTTTCCCTGTGTTCTGGATGTTAAAAGAGGCAAATGAGCTCTTTGTTCTGAGAGCAGATCTGAAACTAGAAAAGATGAAACTTAAAGATTTGACCCACCTCTAATACAAGTCTGGGAATTAGTCAAACTACTCACCGTCCTTACTTTTCAGGAGACCACTTCTACAAGAGAAACAAAAACTTGCCACCCCAGGAACAGATG .....................................................(((((.((((..(((((....)))))..))))))))).......(((((...........)))))........(((((.(((((((((((((((((((..((((.(((..(((((...........((.((((.....)))).))..)))))..)))...))))..)))))..))))))...)))))))))))))................................................................................................................................................. ..................................................51........................................................................................................................................................................................................253.......................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................................................AAAGATGAAACTTAAgcaa.................................................................................................................... | 19 | gcaa | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ATAAGCTCATGCCTTCTTTGTTCTTA...................................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................CTTTGTTCTTAAATACCTCTGTAAAA....................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................GCTCTTCAGAATAAGCTCATGCCTTC................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................TACTCACCGTCCTTACTTTTCAGGAGA.............................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GCCTTCTTTGTTCTTAAATACCTCTGTAAA........................................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................CATGCCTTCTTTGTTCTTAAATACCT............................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TCTTTGTTCTTAAATACCTCTGTAAAA....................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................ACAAGAGAAACAAAAgtgc....................... | 19 | gtgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................TGCCTTCTTTGTTCTTAAATACCTCTGTA.......................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TAAGCCCTGTGCTCTTCAGAATAAGCTCA....................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................AAGCCCTGTGCTCTTCAGAATAAGCTCA....................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................................................................................AGGAGACCACTTCTACAAGAGAAACA.......................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................AGCCCTGTGCTCTTCAGAATAAGCTCA....................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GCAAAGTTTAAGCCCTGTGCTCTTCA.................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................ATAAGCTCATGCCTTCTTTGTTCTTAAA.................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |