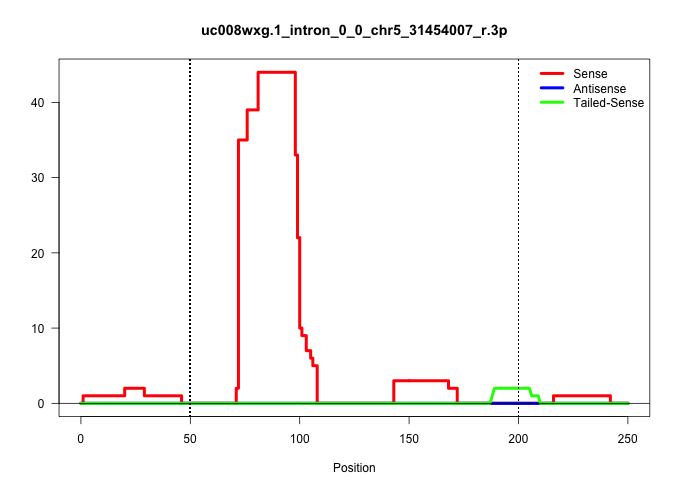
| Gene: Gtf3c2 | ID: uc008wxg.1_intron_0_0_chr5_31454007_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(15) TESTES |
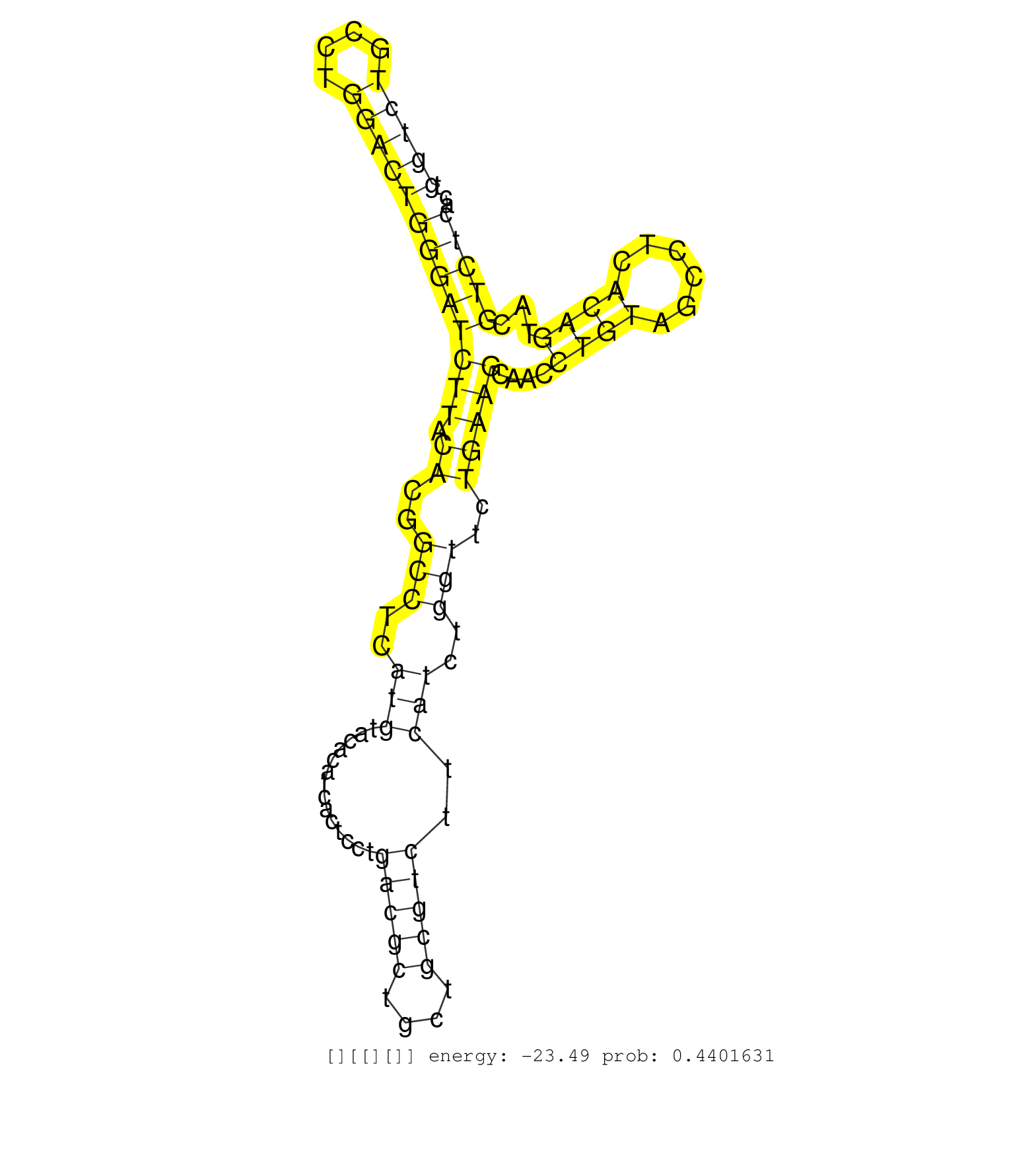
| CTAAATGATCCATAGCTAACTAAAGCTCCAAGGGCACAAGTGCAGAGATGTGGCTCCCTGGGCTCCCTGGTCTGCCTGGACTGGGATCTTACACGGCCTCATGTACACATCACTCCTGACGCTGCTGCGTCTTCATCTGGTTCTGAAGCAACCTGTAGCCTCACAGTACGTCTCACAGACTTCCCCTTCTTTCCTGACAGGGTGTTTGGACCAACCTACAAATAAAAGGTGGGGGTGGGCTTAGCTCACC ....................................................................(((((....)))))((((((((.((..(((..(((..............(((((....)))))..)))..)))..)))))....((((......))))...)))))............................................................................ ...................................................................68..........................................................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................TGCCTGGACTGGGATCTTACACGGCCTC...................................................................................................................................................... | 28 | 1 | 12.00 | 12.00 | 4.00 | 2.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGCCTGGACTGGGATCTTACACGGCCT....................................................................................................................................................... | 27 | 1 | 11.00 | 11.00 | 2.00 | 5.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................TGCCTGGACTGGGATCTTACACGGCC........................................................................................................................................................ | 26 | 1 | 9.00 | 9.00 | 3.00 | 2.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGGGATCTTACACGGCCTCATGTACAC.............................................................................................................................................. | 27 | 1 | 5.00 | 5.00 | 1.00 | - | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TGGACTGGGATCTTACACGGCCTCATG................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................CTGCCTGGACTGGGATCTTACACGGCC........................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGAAGCAACCTGTAGCCTCACAGTACGTC.............................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................TGAAGCAACCTGTAGCCTCACAGTA.................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGCCTGGACTGGGATCTTACACG........................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................TAAAGCTCCAAGGGCACAAGTGCAGA............................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TAAATGATCCATAGCTAACTAAAGCTCC............................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................TACAAATAAAAGGTGGGGGTGGGCTT........ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................................................................TTTCCTGACAGGGTGTTaaca........................................ | 21 | aaca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................TGCCTGGACTGGGATCTTACACGGCCTCA..................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................TGGACTGGGATCTTACACGGCCTCATGTAC................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................CTTTCCTGACAGGGTcct............................................ | 18 | cct | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................TGGACTGGGATCTTACACGGCCTCATGTA................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTAAATGATCCATAGCTAACTAAAGCTCCAAGGGCACAAGTGCAGAGATGTGGCTCCCTGGGCTCCCTGGTCTGCCTGGACTGGGATCTTACACGGCCTCATGTACACATCACTCCTGACGCTGCTGCGTCTTCATCTGGTTCTGAAGCAACCTGTAGCCTCACAGTACGTCTCACAGACTTCCCCTTCTTTCCTGACAGGGTGTTTGGACCAACCTACAAATAAAAGGTGGGGGTGGGCTTAGCTCACC ....................................................................(((((....)))))((((((((.((..(((..(((..............(((((....)))))..)))..)))..)))))....((((......))))...)))))............................................................................ ...................................................................68..........................................................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................ATGTACACATCACTCaagt....................................................................................................................................... | 19 | aagt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TCTTCATCTGGTTCgttt........................................................................................................... | 18 | gttt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |