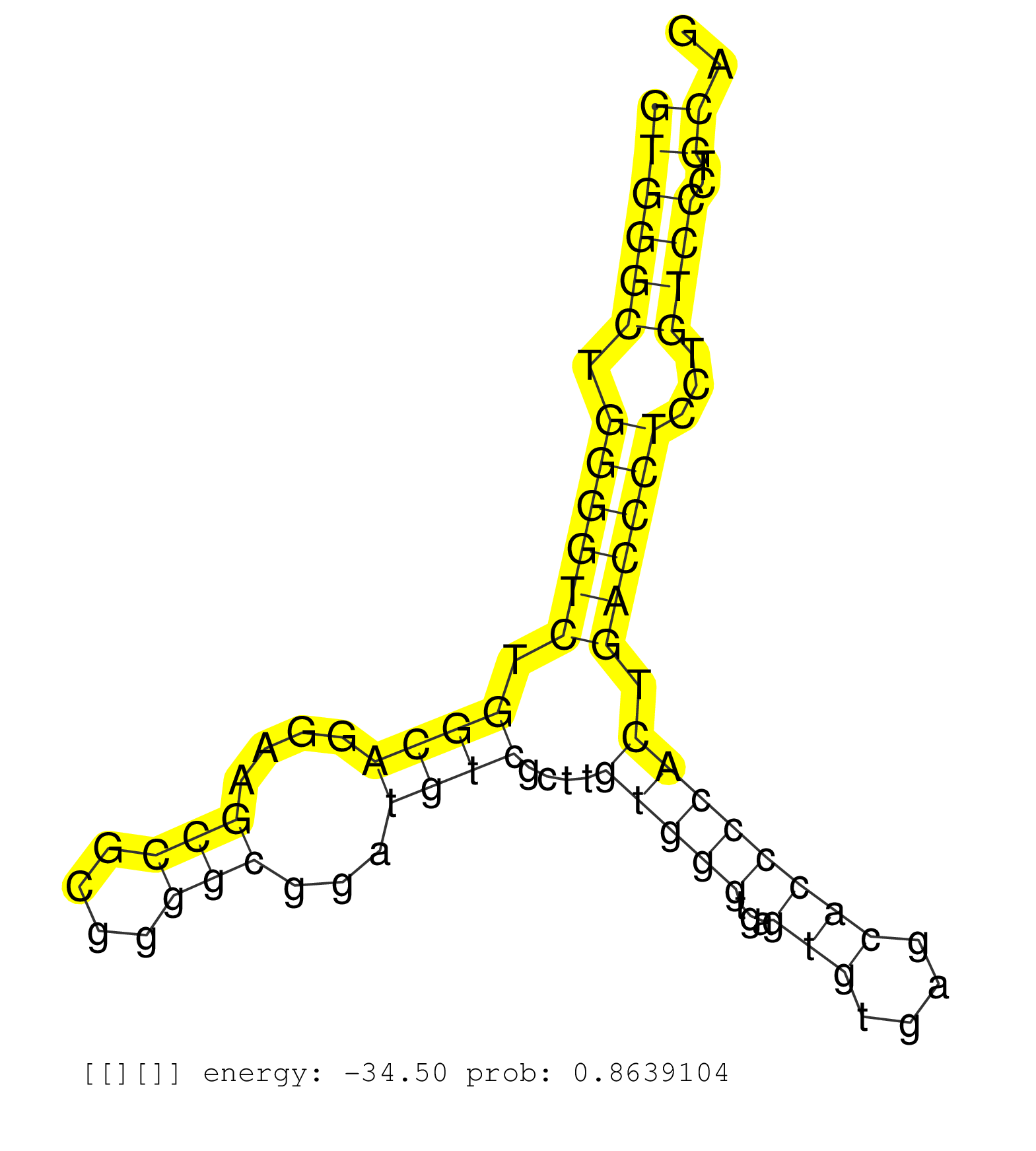

| Gene: Cad | ID: uc008wwz.1_intron_19_0_chr5_31370961_f | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(14) TESTES |
| ATACCATCGGTATCAGCCAGCCTCAGTGGCGAGAGCTCAGTGACCTTGAGGTGGGCTGGGGTCTGGCAGGAAGCCGCGGGGCGGATGTCGCTTGTGGGTGAGTGTGAGCACCCCACTGACCCTCCTGTCCCTGCAGTCTGCTCGCCAGTTCTGCCACACGGTCGGGTACCCCTGCGTAGTGCGCCC ..................................................((((((.((((((.((((....(((....)))...))))....(((((...(((....)))))))).))))))...))))..)).................................................... ..................................................51...................................................................................136................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................ACTGACCCTCCTGTCCCTGCAG.................................................. | 22 | 1 | 9.00 | 9.00 | 3.00 | - | 3.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................GTGGCGAGAGCTCAGTGACCTTGAG........................................................................................................................................ | 25 | 1 | 7.00 | 7.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ACTGACCCTCCTGTCCCTGCAGa................................................. | 23 | a | 3.00 | 9.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCTGGGGTCTGGCAGGAAGCCGC............................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - |
| ..................................................................................................................ACTGACCCTCCTGTCCCTGC.................................................... | 20 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ACTGACCCTCCTGTCCCTGCAGT................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCTGGGGTCTGGCAGGAAG................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................CCCCACTGACCCTCCTGTCCCTGCAGaaa............................................... | 29 | aaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ATACCATCGGTATCAGCCAGCCTCAGTGGCGAGAGCTCAGTGACCTTGAGGTGGGCTGGGGTCTGGCAGGAAGCCGCGGGGCGGATGTCGCTTGTGGGTGAGTGTGAGCACCCCACTGACCCTCCTGTCCCTGCAGTCTGCTCGCCAGTTCTGCCACACGGTCGGGTACCCCTGCGTAGTGCGCCC ..................................................((((((.((((((.((((....(((....)))...))))....(((((...(((....)))))))).))))))...))))..)).................................................... ..................................................51...................................................................................136................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................TCGCCAGTTCTGCtgag................................ | 17 | tgag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................GTCCCTGCAGTCTGCTCGCC........................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................GGTCTGGCAGGAAGC................................................................................................................ | 15 | 12 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 |