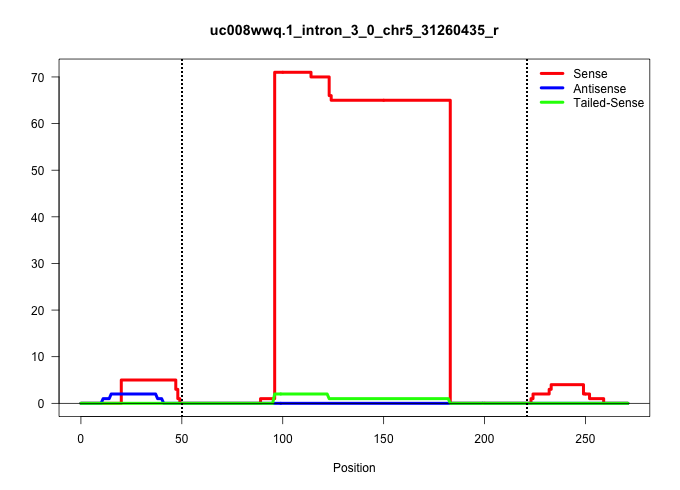
| Gene: Preb | ID: uc008wwq.1_intron_3_0_chr5_31260435_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(11) TESTES |
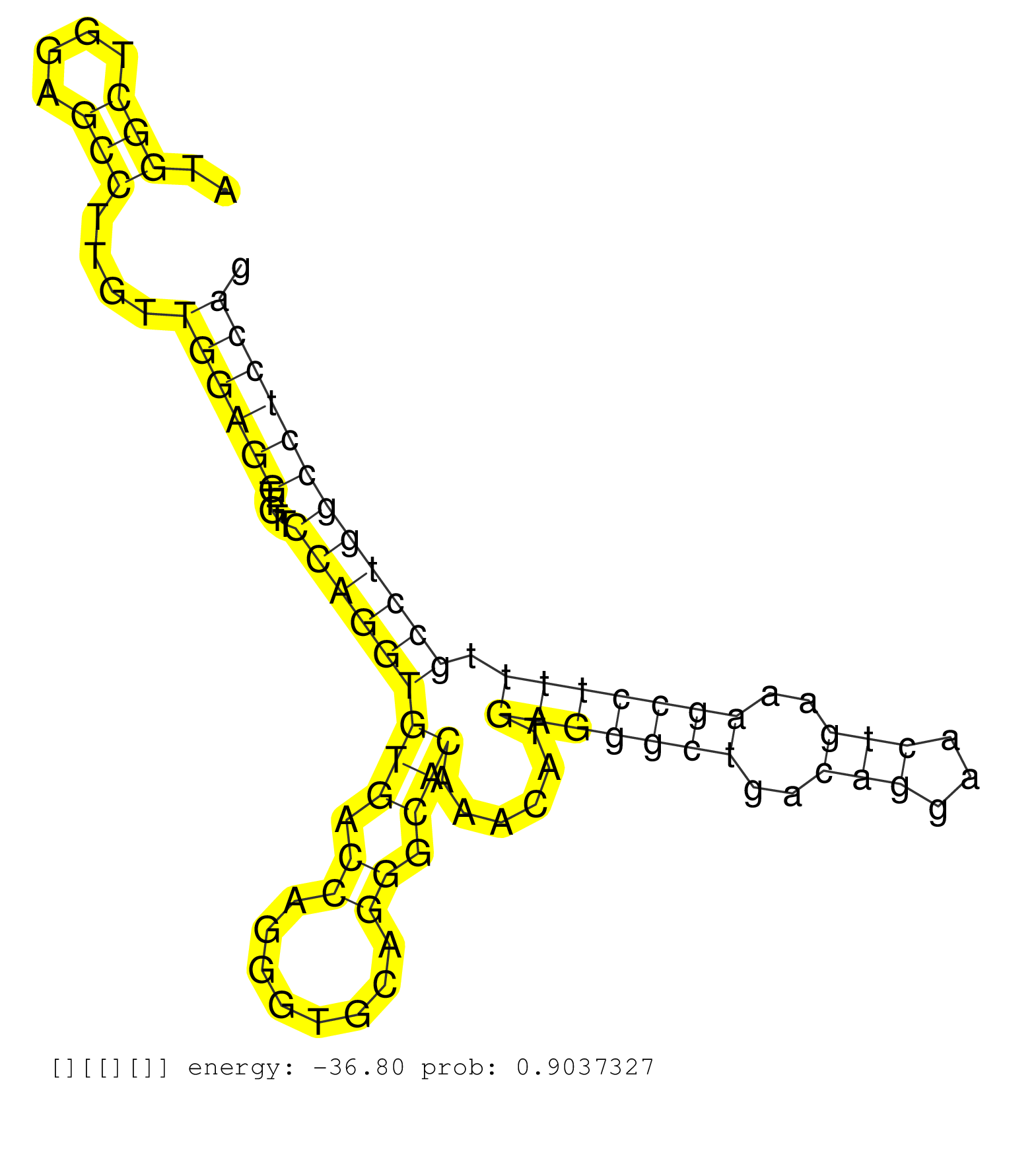
| GAGAATGGACCAGCCTCTTCTAATACACCATACCGCTACCAGGCCTGCAGGTGTGCAGGTCACAGGGTGGCTGGCAGGCACACCTCAGCTGGGAAATGGGAAAAGACAGAAGGAGCTGGCCATGGCTGGAGCCTTGTTGGAGGTTGTTCCAGGTGTGACCAGGGTGCAGGGCACAAACATGAGGGCTGACAGGAACTGAAAGCCTTTTGCCTGGCCTCCAGGTTTGGGCAGGTTCCAGATCAGCTCGGTGGGCTGCGACTCTTCACAGTGC ...........................................................................................................................(((....)))....((((((.....(((((((((.((........)).)))......(((((((..(((...)))..))))))).))))))))))))................................................... .........................................................................................................................122................................................................................................221................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................TGGGAAAAGACAGAAGGAGCTGGCCATGGCTGGAGCCTTGTTGGAGGTTGTT........................................................................................................................... | 52 | 1 | 68.00 | 68.00 | 68.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGGGAAAAGACAGAAGGAGCTGGCCAT.................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................TGGGAAATGGGAAAAGACAGAAGGA............................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - |
| ....................TAATACACCATACCGCTACCAGGCCTGC............................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 |
| ....................TAATACACCATACCGCTACCAGGCCTG................................................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................................TTCCAGATCAGCTCGGTGGG................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................TGGGAAAAGACAGAAGGAGCTGGtcat.................................................................................................................................................... | 27 | tcat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TTGGGCAGGTTCCAGATCAGCTCGGT...................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................GGCACAAACATGAGGGCTGACAGGAACTGAA....................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................TGGGAAAAGACAGAAGGAGCTGGCCATG................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................TAATACACCATACCGCTACCAGGCCTGCA.............................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TGGGCAGGTTCCAGATCAGCTCGGT...................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................TCCAGATCAGCTCGGTGGGCTGCGAC............ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| GAGAATGGACCAGCCTCTTCTAATACACCATACCGCTACCAGGCCTGCAGGTGTGCAGGTCACAGGGTGGCTGGCAGGCACACCTCAGCTGGGAAATGGGAAAAGACAGAAGGAGCTGGCCATGGCTGGAGCCTTGTTGGAGGTTGTTCCAGGTGTGACCAGGGTGCAGGGCACAAACATGAGGGCTGACAGGAACTGAAAGCCTTTTGCCTGGCCTCCAGGTTTGGGCAGGTTCCAGATCAGCTCGGTGGGCTGCGACTCTTCACAGTGC ...........................................................................................................................(((....)))....((((((.....(((((((((.((........)).)))......(((((((..(((...)))..))))))).))))))))))))................................................... .........................................................................................................................122................................................................................................221................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........AGCCTCTTCTAATACACCATACCGCTA......................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............TCTTCTAATACACCATACCGCTACCA...................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |