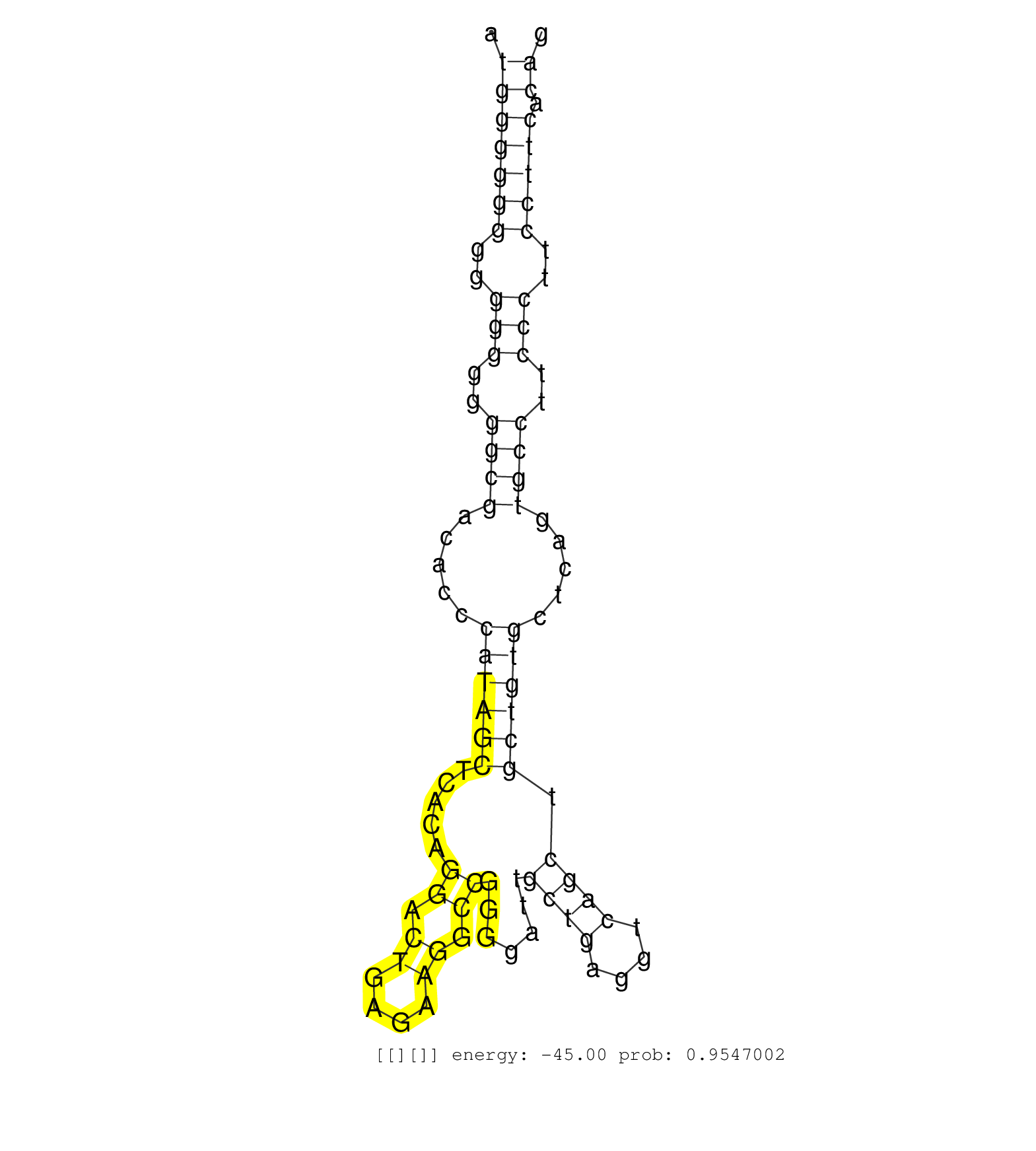
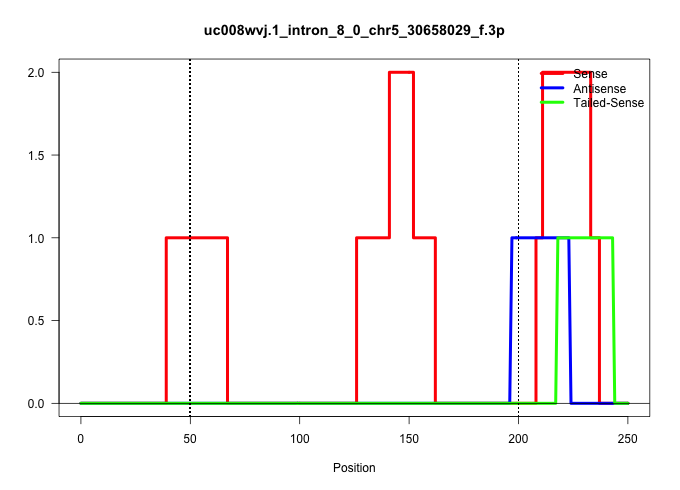
| Gene: Gm1060 | ID: uc008wvj.1_intron_8_0_chr5_30658029_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(1) PIWI.mut |
(7) TESTES |
| AGGGATGACATCAGAGCTAAAGTCATAGGAAGGTCACTCTGCTAGAGAGCCAGCCAGAACAGGGTGTTCAGAAAGGTGGCAGAAGACAGTCCCTCGGAAGATGGGGGGGGGGGGGGGCGACACCCATAGCTCACAGGACTGAGAAGGCCGGGGATTGCTGAGGTCAGCTGCTGTGCTCAGTGCCTTCCCTTCCTTCACAGACACTTCATCATCATCGATGAGGAGAAGTTTCGGGAGATTTGGCTGATGA .....................................................................................................(((((((..(((..((((.....((((((.....((.((....)).)).......((((....)))).)))))).....))))..)))..))))).))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................TAGCTCACAGGACTGAGAAGGCCGGG.................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| .......................................TGCTAGAGAGCCAGCCAGAACAGGGTGT....................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................................................................................................................TCATCATCGATGAGGAGAAGTTTCG................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| .......................................TGCTAGAGAGCCAGCCAGAACAGGGcg........................................................................................................................................................................................ | 27 | cg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................................................TCATCGATGAGGAGAAGTTTCGGGAG............. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TGAGGAGAAGTTTCGGGAGATTTGGa...... | 26 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................AGAAGGCCGGGGATTGCTGAG........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................................CTCACAGGACTGAGAcgt....................................................................................................... | 18 | cgt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................ACAGGACTGAGAAGGC...................................................................................................... | 16 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - | - | - | - |
| AGGGATGACATCAGAGCTAAAGTCATAGGAAGGTCACTCTGCTAGAGAGCCAGCCAGAACAGGGTGTTCAGAAAGGTGGCAGAAGACAGTCCCTCGGAAGATGGGGGGGGGGGGGGGCGACACCCATAGCTCACAGGACTGAGAAGGCCGGGGATTGCTGAGGTCAGCTGCTGTGCTCAGTGCCTTCCCTTCCTTCACAGACACTTCATCATCATCGATGAGGAGAAGTTTCGGGAGATTTGGCTGATGA .....................................................................................................(((((((..(((..((((.....((((((.....((.((....)).)).......((((....)))).)))))).....))))..)))..))))).))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................CAGACACTTCATCATCATCGATGAGGA.......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |