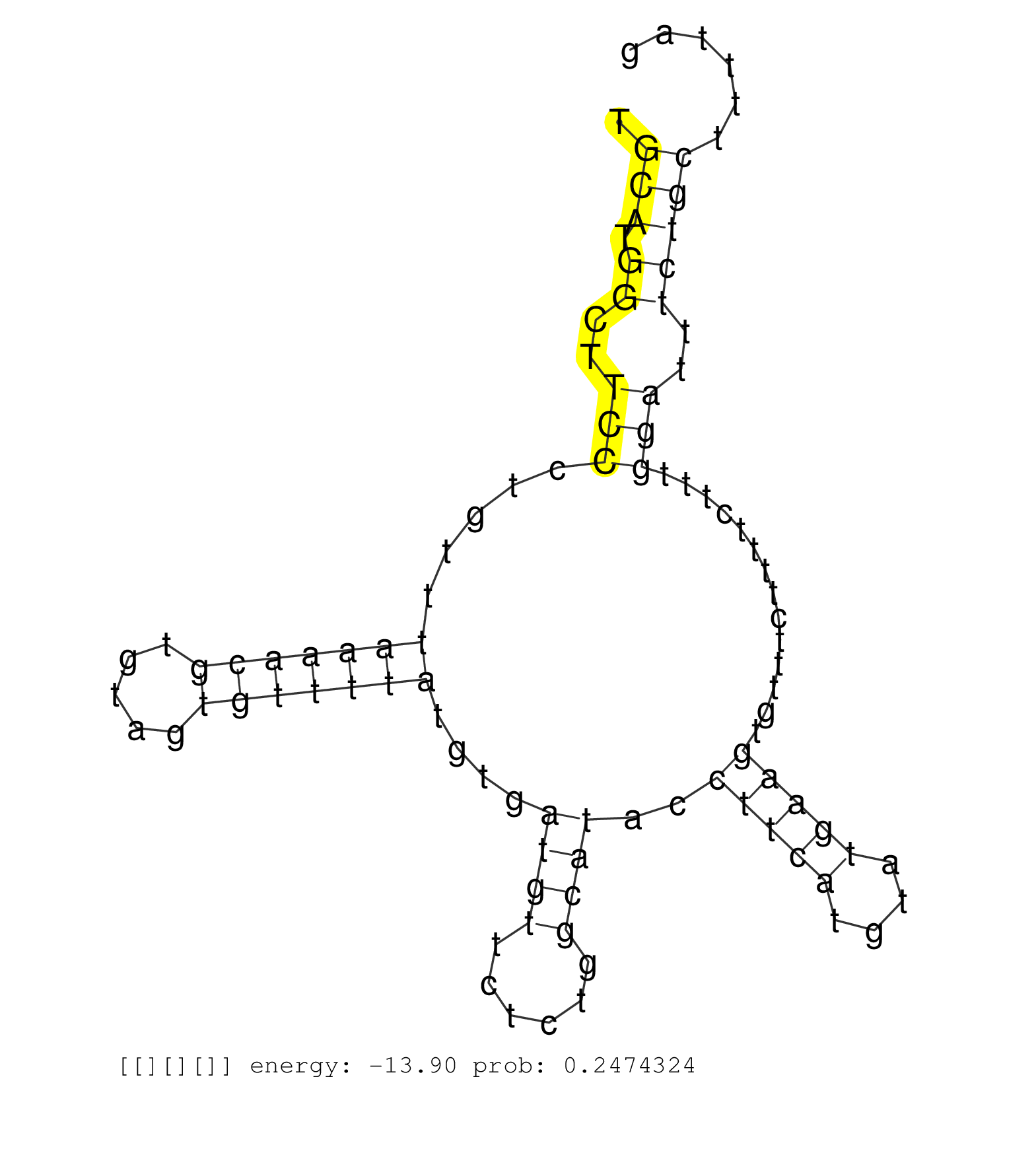

| Gene: Hadhb | ID: uc008wve.1_intron_4_0_chr5_30495109_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| GAAAAGGAAAGACTTGTCTTTGTGTTTTTGTGGAGATTAGGATTGAACCGAGGGCCTTGCATGTACTAAATGAGCACTGTCCTCTGGCCTGTACCTCCAGTGCATGGCTTCCCTGTTTAAAACGTGTAGTGTTTTATGTGATGTTCTCTGGCATACCTTCATGTATGAAGTGTTTCTTTTCTTTGGATTTCTGCTTTTAGGGGTTTGTTGCATCGGACCAATATTCCAAAGGATGTTGTTGATTATATCA .....................................................................................................(((.((..(((.....(((((((.....)))))))....((((......))))..(((((....)))))..............)))..)))))........................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................ATGTACTAAATGAGCACTGTCCTCTGGCCTGTACCTCCAGTGCATGGCTTCC.......................................................................................................................................... | 52 | 1 | 21.00 | 21.00 | 21.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................GCATCGGACCAATATTCCAAAGGATG............... | 26 | 2 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TTGCATCGGACCAATATTCCAAAGGATGT.............. | 29 | 2 | 1.50 | 1.50 | - | - | 1.00 | - | - | - | - | - | - | 0.50 | - |
| ........................................................................................................................AACGTGTAGTGTTTggcc................................................................................................................ | 18 | ggcc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................GTTGCATCGGACCAATATTCCAAAGGATGT.............. | 30 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - |
| .........................................................................................................................................................................................................GGTTTGTTGCATCGGAC................................ | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ................................................................................................................................................................................................................TGCATCGGACCAATATTCCAAAGGATG............... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ................................................................................................................................................................................................................TGCATCGGACCAATATTCCAAAGGATGT.............. | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ............................................................................................................................................................................................................................ATATTCCAAAGGATGTTGTTGATTATATC. | 29 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | 0.33 |
| GAAAAGGAAAGACTTGTCTTTGTGTTTTTGTGGAGATTAGGATTGAACCGAGGGCCTTGCATGTACTAAATGAGCACTGTCCTCTGGCCTGTACCTCCAGTGCATGGCTTCCCTGTTTAAAACGTGTAGTGTTTTATGTGATGTTCTCTGGCATACCTTCATGTATGAAGTGTTTCTTTTCTTTGGATTTCTGCTTTTAGGGGTTTGTTGCATCGGACCAATATTCCAAAGGATGTTGTTGATTATATCA .....................................................................................................(((.((..(((.....(((((((.....)))))))....((((......))))..(((((....)))))..............)))..)))))........................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................TCTCTGGCATACCTttt............................................................................................ | 17 | ttt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |