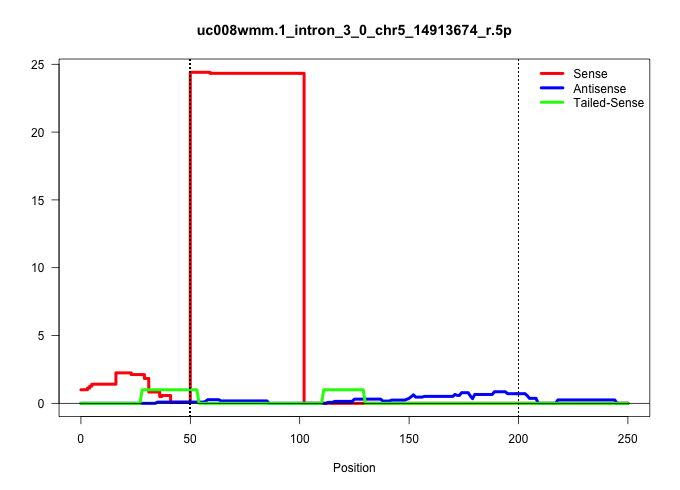
| Gene: ENSMUSG00000033219 | ID: uc008wmm.1_intron_3_0_chr5_14913674_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(23) TESTES |
| TCCATCTGAGAGGAATGAAGGACGAAGGAGGGGGACTTGGCGAATGTGGAGTAAGTCCTGGGAAGTGGATGTTGAGCTTCCTGGTAGGGGTGGCCTGAGGTGGCCTTTCCAGTAATGTGGAACTGGAGACTCTGGGTAGTGAAAACATTTCCATGGTTATCACAATTCTTGAACATCAAGGATTTCAGAACAGTACTTTTCCATCCCCTAAGGCAGGCGTTGGCAAGGTCAAAGCTGTTATGCTATGAAC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTCCTGGGAAGTGGATGTTGAGCTTCCTGGTAGGGGTGGCCTGAGGTG.................................................................................................................................................... | 52 | 3 | 24.33 | 24.33 | 24.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................GTAATGTGGAACTGGtggt........................................................................................................................ | 19 | tggt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................AGGGGGACTTGGCGAATGTGGAtggc.................................................................................................................................................................................................... | 26 | tggc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GAAGGACGAAGGAGGGGGACTTGGC................................................................................................................................................................................................................. | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GAAGGACGAAGGAGGGGGAC...................................................................................................................................................................................................................... | 20 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CTGAGAGGAATGAAGGACGAAGGA............................................................................................................................................................................................................................. | 24 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - |
| ....TCTGAGAGGAATGAAGGACGAAGGA............................................................................................................................................................................................................................. | 25 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - |
| ...ATCTGAGAGGAATGAAGGAC................................................................................................................................................................................................................................... | 20 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TGGCGAATGTGGAGTAAGTCCT............................................................................................................................................................................................... | 22 | 13 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 | - | - |
| TCCATCTGAGAGGAATGAAGGACGAAGGAGGGGGACTTGGCGAATGTGGAGTAAGTCCTGGGAAGTGGATGTTGAGCTTCCTGGTAGGGGTGGCCTGAGGTGGCCTTTCCAGTAATGTGGAACTGGAGACTCTGGGTAGTGAAAACATTTCCATGGTTATCACAATTCTTGAACATCAAGGATTTCAGAACAGTACTTTTCCATCCCCTAAGGCAGGCGTTGGCAAGGTCAAAGCTGTTATGCTATGAAC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................CGAATGTGGAGTAAGTCCTGGgc............................................................................................................................................................................................. | 23 | gc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CTAAGGCAGGCGTTGGCAAGGTCAAAa................. | 27 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TTATCACAATTCTTGAACATCAtgga........................................................................ | 26 | tgga | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ATTTCCATGGTTATCACAATTCTTGAAa............................................................................. | 28 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................CGAATGTGGAGTAAGTCCTGGGgc............................................................................................................................................................................................ | 24 | gc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................GTTGGCAAGGTCAAAGCTGTTATGCTA..... | 27 | 8 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................ATCAAGGATTTCAGAACAGTACTTTTCCATC............................................. | 31 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................ACAGTACTTTTCCATCCCCT......................................... | 20 | 10 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | 0.10 | - | - | - | - | - | - | - |
| .............................................................................................................................GAGACTCTGGGTAGTGAAAACATTTCCA................................................................................................. | 28 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................GATTTCAGAACAGTACTTTTCCATCCCCT......................................... | 29 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................GATTTCAGAACAGTACTTTTCCAT.............................................. | 24 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - |
| ...........................................................................................................................................................................AACATCAAGGATTTCAGAACAGTA....................................................... | 24 | 7 | 0.14 | 0.14 | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................ATGGTTATCACAATTCTTGAACATCAA....................................................................... | 27 | 15 | 0.13 | 0.13 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.13 | - | - | - | - |
| .......................................................................................................................................................CATGGTTATCACAATTCTTGAACATCA........................................................................ | 27 | 16 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | 0.06 |
| ..........................................................TGGGAAGTGGATGTTGAGCTTCCTGGTA.................................................................................................................................................................... | 28 | 17 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................CTTGGCGAATGTGGAGTAAGTCCTGGGAA.......................................................................................................................................................................................... | 29 | 11 | 0.09 | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.09 | - | - | - |
| ..............................................................................................................................................AAACATTTCCATGGTTATCACAATTCTTGA.............................................................................. | 30 | 14 | 0.07 | 0.07 | - | - | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................GTGGAACTGGAGACTCTGGGTA................................................................................................................ | 22 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - | - | - | - | - | - | - |
| .................................................................................................................AATGTGGAACTGGAGACTCTGGGTA................................................................................................................ | 25 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - |
| ......................................................................................................................................................CCATGGTTATCACAATTCTTGAACATCAA....................................................................... | 29 | 15 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TCCATGGTTATCACAATTCTTGAACATCA........................................................................ | 29 | 16 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - |
| .........................................................CTGGGAAGTGGATGTTGAGCTTCCTGGTA.................................................................................................................................................................... | 29 | 17 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TATCACAATTCTTGAACATCA........................................................................ | 21 | 19 | 0.05 | 0.05 | - | - | - | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |