
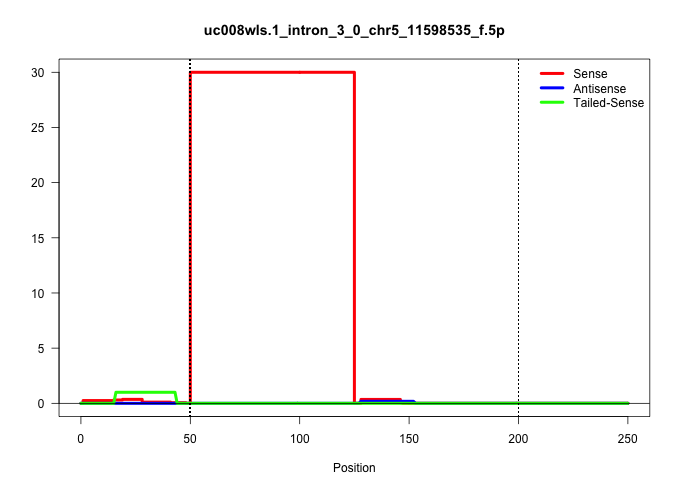
| Gene: EG623898 | ID: uc008wls.1_intron_3_0_chr5_11598535_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(1) PIWI.ip |
(1) TDRD1.ip |
(11) TESTES |
| GAATGGACAAACAGGAGCAGACCAGCAACCATGAGACCCAACAACTTCAGGTAGGGACAAGCAGCTCACTCAAGCTAATACTGTCTGACACCATTTGCCCCATTGGATTCATCTTTGCTTTCCCCTACCACCTTTCTAATCCACACTCCCAAACAGTCACCCACCTCTTGGCATGTAGTTGTTCATTGCCCTTTTTATATACACATATTCTGGGAAGTTAGCCTCTTGTGAGAAACTAAATCATTGGCAA ..................................................((((((..((((((.....(((.((.......)).)))..(((..........)))........))))))..)))))).......................................................................................................................... ..................................................51.................................................................................................150.................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGGACAAGCAGCTCACTCAAGCTAATACTGTCTGACACCATTTGCCCCA.................................................................................................................................................... | 52 | 4 | 23.00 | 23.00 | 23.00 | - | - | - | - | - | - | - | - | - | - |
| ................GCAGACCAGCAACCATGAGACCCAACAt.............................................................................................................................................................................................................. | 28 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .AATGGACAAACAGGAGCAGACCAGCAA.............................................................................................................................................................................................................................. | 27 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - | - | - | - | - | - | - |
| ...................GACCAGCAACCATGAGACCCAACAACTTC.......................................................................................................................................................................................................... | 29 | 18 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | 0.06 |
| ...............AGCAGACCAGCAACCATGAGACCCAA................................................................................................................................................................................................................. | 26 | 19 | 0.05 | 0.05 | - | 0.05 | - | - | - | - | - | - | - | - | - |
| GAATGGACAAACAGGAGCAGACCAGCAACCATGAGACCCAACAACTTCAGGTAGGGACAAGCAGCTCACTCAAGCTAATACTGTCTGACACCATTTGCCCCATTGGATTCATCTTTGCTTTCCCCTACCACCTTTCTAATCCACACTCCCAAACAGTCACCCACCTCTTGGCATGTAGTTGTTCATTGCCCTTTTTATATACACATATTCTGGGAAGTTAGCCTCTTGTGAGAAACTAAATCATTGGCAA ..................................................((((((..((((((.....(((.((.......)).)))..(((..........)))........))))))..)))))).......................................................................................................................... ..................................................51.................................................................................................150.................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................CACCTTTCTAATCCACACTCCCAacgt................................................................................................... | 27 | acgt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TAATCCACACTCCCActca................................................................................................... | 19 | ctca | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................CATCTTTGCTTTCCCCTACCACCTTTCTAt................................................................................................................ | 30 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................CACCTTTCTAATCCACACTCtat...................................................................................................... | 23 | tat | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................CACCTTTCTAATCCACACTCCCAAA................................................................................................. | 25 | 11 | 0.18 | 0.18 | - | - | - | - | - | - | - | 0.09 | 0.09 | - | - |
| ....................................................................................................................................TTTCTAATCCACACTCCCAAACAGTCA........................................................................................... | 27 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | 0.10 | - | - | - | - |
| ............................................................................................................................................................................................................................GCCTCTTGTGAGAAACTAAATC........ | 22 | 17 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | 0.06 | - |