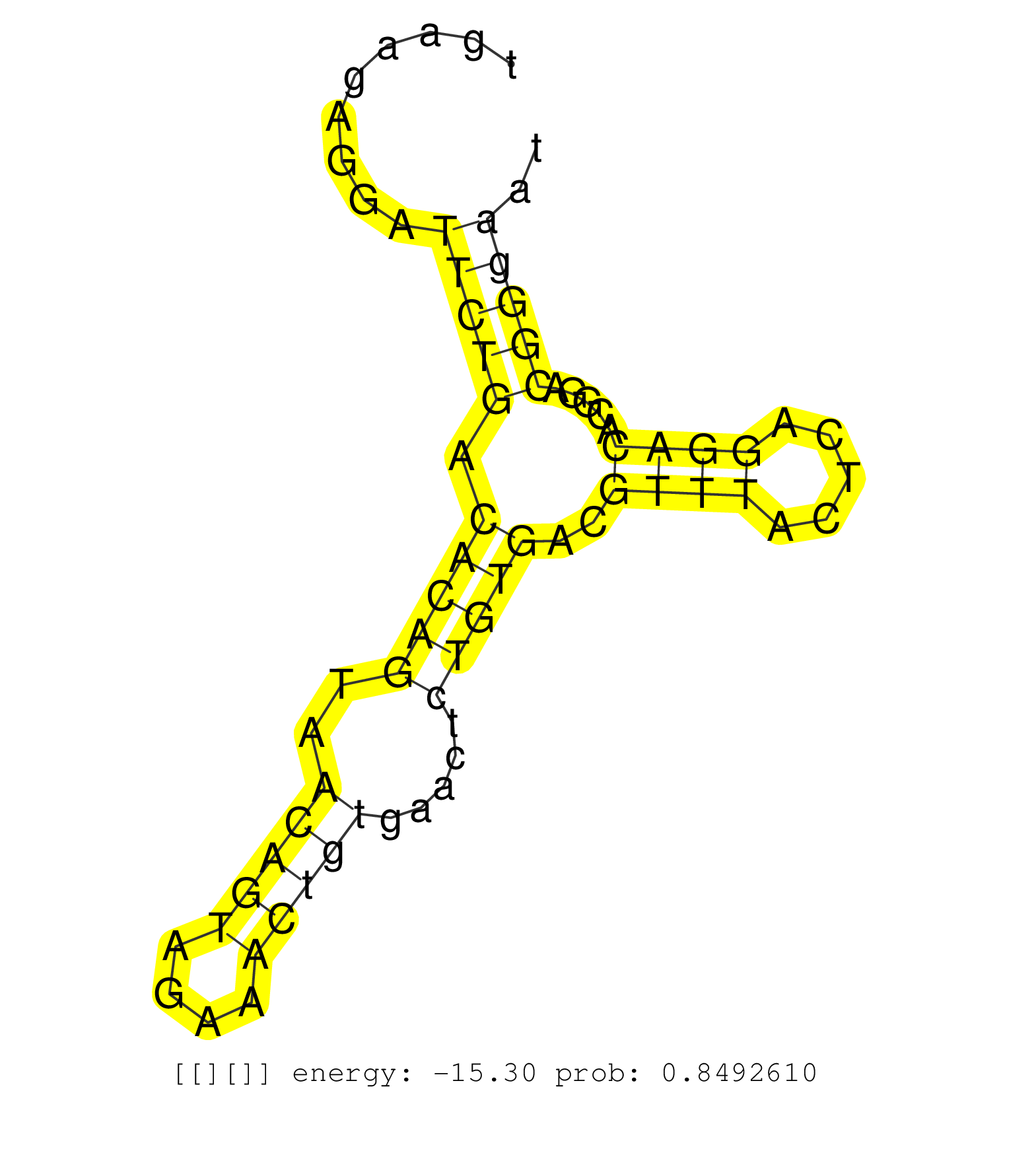

| Gene: Dmtf1 | ID: uc008wlc.1_intron_3_0_chr5_9121841_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(19) TESTES |
| GTATAAGTCCTTACATAGTCTCTTCTTATACAGATTTGAGTATGAGCACAGTTGAAGAGGATTCTGACACAGTAACAGTAGAAACTGTGAACTCTGTGACGTTTACTCAGGACACGGACGGGAATCTCATTCTTCATTGCCCTCAGAATGGTAGGAGAATTTGCTAATTACTGACACTGGGGATTGATCATTGCACCCTTGCACCCTTTATCCAACACCCACAAATTAGTCTAATGTGCTTGAAAAGGCC .............................................................(((((.(((((..(((((....))))).....)))))..((((.....)))).....)))))............................................................................................................................... ....................................................53......................................................................125........................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................GTTTACTCAGGACACGGA.................................................................................................................................... | 18 | 1 | 12.00 | 12.00 | - | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................AGATTTGAGTATGAGCACAGTTGAAG................................................................................................................................................................................................. | 26 | 1 | 10.00 | 10.00 | 4.00 | - | - | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - |
| ...............................AGATTTGAGTATGAGCACAGTTGAAGAG............................................................................................................................................................................................... | 28 | 1 | 5.00 | 5.00 | - | - | - | - | - | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................TGTGACGTTTACTCAGGACACGGACGG................................................................................................................................. | 27 | 1 | 5.00 | 5.00 | 2.00 | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGAGCACAGTTGAAGAGGATTCTGACAC.................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGACGTTTACTCAGGACACGGACGGG................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TGAGTATGAGCACAGTTGAAGAGGATTC.......................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGGATTCTGACACAGTAACAGTAGAAAC..................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................AACTGTGAACTCTGTGACGTTTA................................................................................................................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TGAAGAGGATTCTGACACAGTAACAGTA.......................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGAGCACAGTTGAAGAGGATTCTGACA..................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................GTATGAGCACAGTTGAAGAGGATTCTG........................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................AGAGGATTCTGACACAGTAACAGTAG......................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TGAAGAGGATTCTGACACAGTAACAGTAG......................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TTTACTCAGGACACGGACGGGAATCT........................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................TGTGAACTCTGTGACGTTTACTCAGGA.......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................TTGAGTATGAGCACAGTTGAAGAGGATT........................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGTAGAAACTGTGAACTCTGTGACGTTTACTC.............................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................TTCTGACACAGTAACAGTAGAAACTGTGAAg.............................................................................................................................................................. | 31 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................AATTACTGACACTGGGGATT................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................TGTGACGTTTACTCAGGACACGGACG.................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................GAAGAGGATTCTGACACAGTAACAGTAGA........................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................AGTATGAGCACAGTTGAAGAGGATTCTtt....................................................................................................................................................................................... | 29 | tt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................ACTCTGTGACGTTTACTCAGGACACGGAC................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TGTGAACTCTGTGACGTTTACTCAGGACACGG..................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................AGATTTGAGTATGAGCACAGTTGA................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................AGATTTGAGTATGAGCACAGTTGAAGAGGA............................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................GAAGAGGATTCTGACACAGTAACAGTAGAA....................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................TGAGTATGAGCACAGTTGAAGAGGATTCTG........................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGGATTCTGACACAGTAACAGTAGAAA...................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGACGTTTACTCAGGACACGGACGGGA............................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TATGAGCACAGTTGAAGAGGATTCTGAC...................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGACGTTTACTCAGGACACGGACGGGtt.............................................................................................................................. | 28 | tt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................GAATTTGCTAATTACTGACACTGGGGATTGATtatt.......................................................... | 36 | tatt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................GCTAATTACTGACACTGGGGATTG................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................TGAAGAGGATTCTGACACAGTAAC.............................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGAGCACAGTTGAAGAGGATTCTGACACA................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TGAAGAGGATTCTGACACAGTAACAGT........................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................TGAGTATGAGCACAGTTGAAGAGGATT........................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TCAGGACACGGACGGGAATCTCATT....................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................TGAAGAGGATTCTGACACAGTAACAGTAGA........................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................AGTATGAGCACAGTTGAAGAGGATTCTG........................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTATAAGTCCTTACATAGTCTCTTCTTATACAGATTTGAGTATGAGCACAGTTGAAGAGGATTCTGACACAGTAACAGTAGAAACTGTGAACTCTGTGACGTTTACTCAGGACACGGACGGGAATCTCATTCTTCATTGCCCTCAGAATGGTAGGAGAATTTGCTAATTACTGACACTGGGGATTGATCATTGCACCCTTGCACCCTTTATCCAACACCCACAAATTAGTCTAATGTGCTTGAAAAGGCC .............................................................(((((.(((((..(((((....))))).....)))))..((((.....)))).....)))))............................................................................................................................... ....................................................53......................................................................125........................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....CCTTACATAGTCTtagg..................................................................................................................................................................................................................................... | 17 | tagg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |