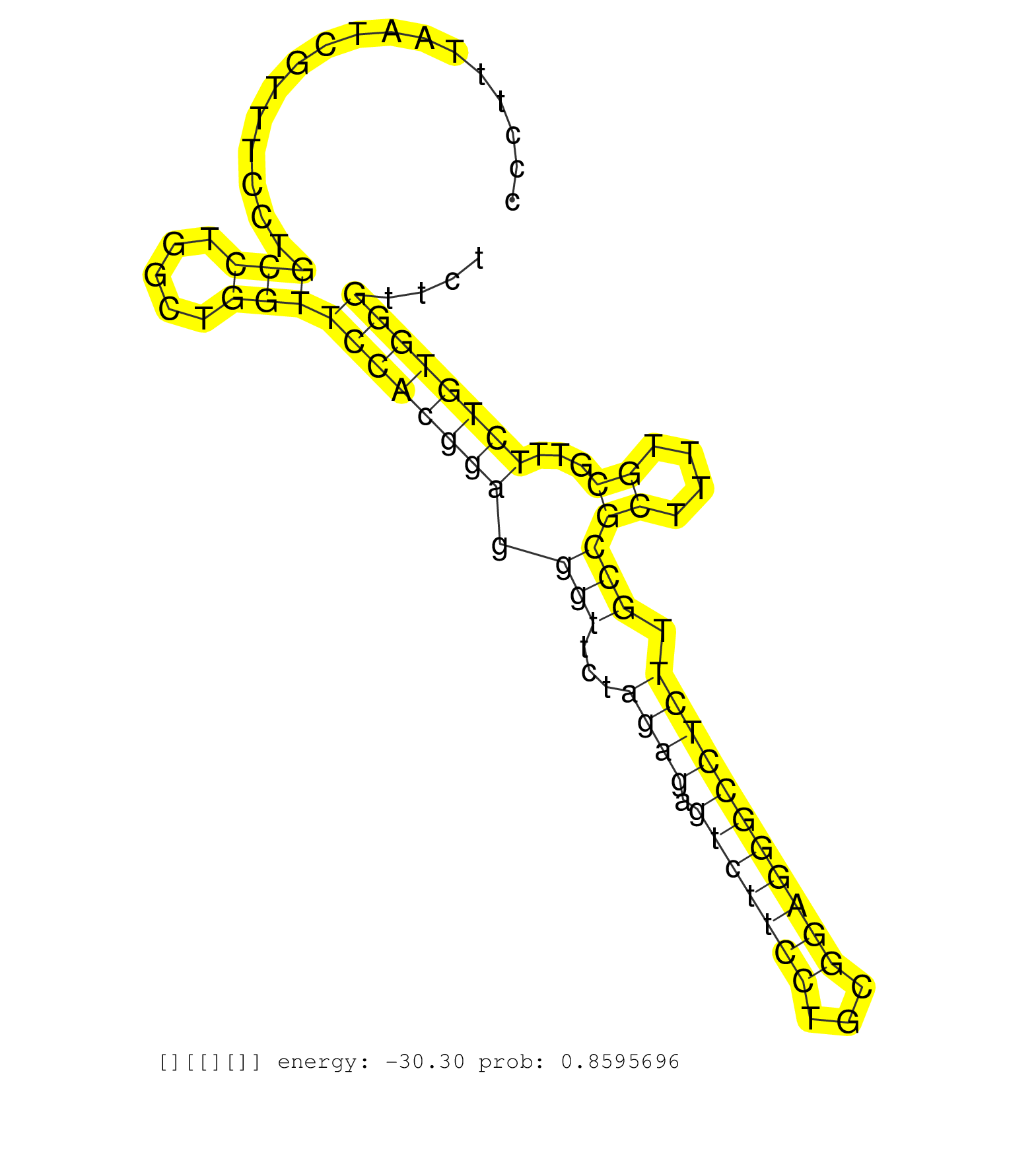

| Gene: Dbf4 | ID: uc008wkc.1_intron_10_0_chr5_8421394_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(13) TESTES |
| TGCCATGAACCTCGAGACCATGAGGATCCACAGCAAAGCACCTCTCCCGGGTATGGAGCCCCTCCCCCGCCGCGGTCCCTTTAATCGTTTCCTGCCTGGCTGGTTCCACGGAGGGTTCTAGAGAGTCTTCCTGCGGAGGGCCTCTTGCCGCTTTTGCGTTTCTGTGGGTTCTCGGCGCCCGCGAGGCGTTCGGTGTTGGTCTCCGTGAGGGACCGACGCTGGCCGGGCCCCTCATGCTCTCCCCGCCTCG .............................................................................................(((.....)))((((((((.(((...((((.(((((((...))))))))))).)))((....))...)))))))).................................................................................. ............................................................................77.............................................................................................172............................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................CCTGCGGAGGGCCTCTTGCCGCTTTTGCGTTTCTGTGGGTTCTCGGCGCCCG..................................................................... | 52 | 1 | 49.00 | 49.00 | 49.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............CGAGACCATGAGGATCCACAGCAAAGCACC................................................................................................................................................................................................................ | 30 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - |
| ..............AGACCATGAGGATCCACAGCAAAGCAC................................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ............CGAGACCATGAGGATCCACAGCAAAGCAa................................................................................................................................................................................................................. | 29 | a | 2.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................GCGTTTCTGTGGGTTCTCGGCG......................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................GCAAAGCACCTCTCCCGG........................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............CGAGACCATGAGGATCCACAGCAAAG.................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................TAATCGTTTCCTGCCTGGCTGGTTCCA.............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....TGAACCTCGAGACCATGAGGATCCACAG......................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............GACCATGAGGATCCACAGCAAAGCACCTC.............................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................CTAGAGAGTCTTCCTGCGGAGGGC............................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................TGGTTCCACGGAGGGTTCTAGAGAGT............................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............CGAGACCATGAGGATCCACAGCAAAGCA.................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............AGACCATGAGGATCCACA.......................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| TGCCATGAACCTCGAGACCATGAGGATCCACAGCAAAGCACCTCTCCCGGGTATGGAGCCCCTCCCCCGCCGCGGTCCCTTTAATCGTTTCCTGCCTGGCTGGTTCCACGGAGGGTTCTAGAGAGTCTTCCTGCGGAGGGCCTCTTGCCGCTTTTGCGTTTCTGTGGGTTCTCGGCGCCCGCGAGGCGTTCGGTGTTGGTCTCCGTGAGGGACCGACGCTGGCCGGGCCCCTCATGCTCTCCCCGCCTCG .............................................................................................(((.....)))((((((((.(((...((((.(((((((...))))))))))).)))((....))...)))))))).................................................................................. ............................................................................77.............................................................................................172............................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................GGAGCCCCTCCCCCGCCGCGGTCCCTT......................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................TGCCGCTTTTGCGTTTCTGTGGGTTCTCG............................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |