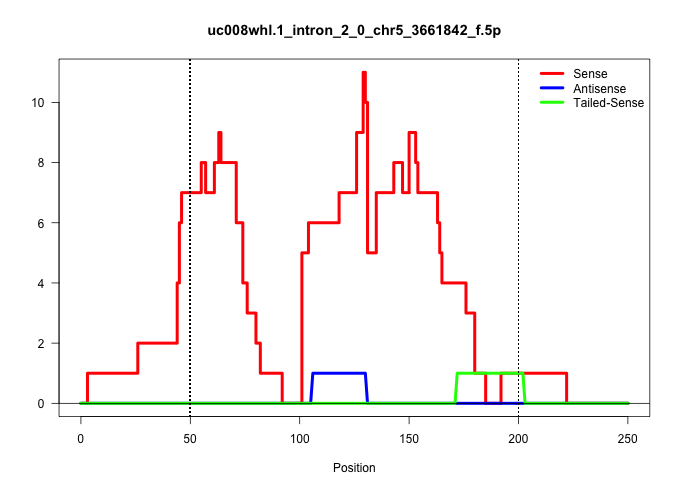
| Gene: 4930511M11Rik | ID: uc008whl.1_intron_2_0_chr5_3661842_f.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(2) PIWI.ip |
(1) TDRD1.ip |
(17) TESTES |
| CTTTAGCTCTGGAGATCATCAACATCTGGTTCATACAGCAGCCATTAAAGGTAACACTGGCAGACTGGATCAGCCCTTCACAGCTTCATCTGGAGACAGGGTACAAGAAGCCGTACAATACTCACATCCAAGGAATAGAGATGGAGCAAATACCTATGCAGTACAAGTTTCTGAGGATGTTACTCCCTCAAATGAGACTGTAAATCAATCCATTCCCTTTGAAAACACGTCTATTCGTAATGGTAAGAGT ....................................................................................................((((........)))).......((((((.((((((....((...(((........))).))....))))))..))))))...................................................................... ................................................................................................97.....................................................................................184................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................TACAAGAAGCCGTACAATACTCACATCCAA....................................................................................................................... | 30 | 1 | 5.00 | 5.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................TACTCACATCCAAGGAATAG................................................................................................................ | 20 | 1 | 4.00 | 4.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TTAAAGGTAACACTGGCAGACTGGATCAGC................................................................................................................................................................................ | 30 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TACCTATGCAGTACAAGTTTCTGAGGATGT...................................................................... | 30 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AAGAAGCCGTACAATACTCACATCCA........................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................TAGAGATGGAGCAAATACCTATGCAGTACA..................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................GAGGATGTTACTCCCTCAAATGAGACTGTAt............................................... | 31 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................ACTGGATCAGCCCTTCACAGCTTCATCTG.............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................AAGGAATAGAGATGGAGCAAATACC................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................AAGGAATAGAGATGGAGCAAATAC................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................AGACTGGATCAGCCCTTCACA........................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TCCAAGGAATAGAGATGGAGC....................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................TGGTTCATACAGCAGCCATTAAAGGTAACAC................................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................AATACCTATGCAGTACAAGTTTCTGAGG.......................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TAAAGGTAACACTGGCAGACTGGATC................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TACTCACATCCAAGGAATAGAGATGGAGCA...................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGCAGTACAAGTTTCTGAGGATGTTACTC................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................ATACAGCAGCCATTAAAGGTAACACTGGCAGA.......................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TGAGACTGTAAATCAATCCATTCCCTTTGA............................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GAGCAAATACCTATGCAGTA....................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TAGAGATGGAGCAAATACCTATGCAGTAC...................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TAGCTCTGGAGATCATCAACATCTGGTTC.......................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................TCCAAGGAATAGAGATGGAGCAAATACCTA.............................................................................................. | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TAAAGGTAACACTGGCAGACTGGATCAGCCC.............................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................ACTGGCAGACTGGATCAGCCCTTCA.......................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................AAAGGTAACACTGGCAGACTGGATC................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| CTTTAGCTCTGGAGATCATCAACATCTGGTTCATACAGCAGCCATTAAAGGTAACACTGGCAGACTGGATCAGCCCTTCACAGCTTCATCTGGAGACAGGGTACAAGAAGCCGTACAATACTCACATCCAAGGAATAGAGATGGAGCAAATACCTATGCAGTACAAGTTTCTGAGGATGTTACTCCCTCAAATGAGACTGTAAATCAATCCATTCCCTTTGAAAACACGTCTATTCGTAATGGTAAGAGT ....................................................................................................((((........)))).......((((((.((((((....((...(((........))).))....))))))..))))))...................................................................... ................................................................................................97.....................................................................................184................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................AGCAAATACCTATGCAattt.......................................................................................... | 20 | attt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................GAAGCCGTACAATACTCACATCCAA....................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |