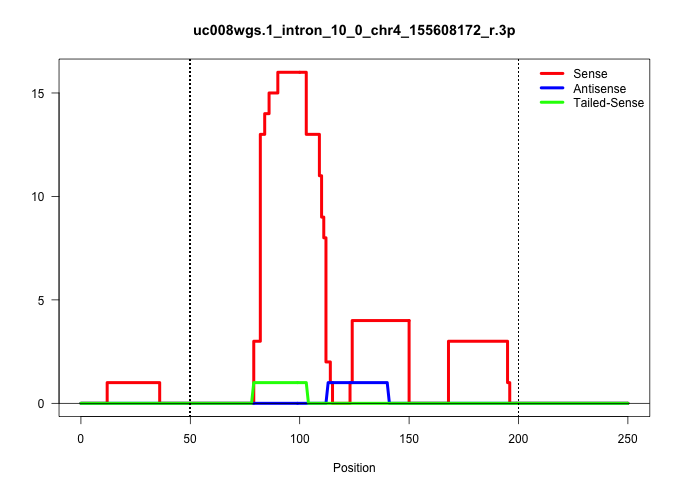
| Gene: Klhl17 | ID: uc008wgs.1_intron_10_0_chr4_155608172_r.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| GTTCCTGGACCGGAGAGAGATCTAGAGCCCTGGAAAGTGGCAGTGAGTGACAGGCTGGTCCTCCATGGGCTTTGGTGCCTGATGACTGAACGAACTCCTGGGCATAGACCGCAGCTGCCTTGGCTGTGCTGGGAGTTCTCAAAGGCGGGTGAGTGGCCCTCTACAGGGTACAAACCCTGACCTAGTCCTGCGGCCTGCAGCCCCGAGGCAGAGCGAGCTCGGCCCCGGCAGGCCCGGCCTGCAGCCCCGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................TGACTGAACGAACTCCTGGGCATAGACCGC.......................................................................................................................................... | 30 | 1 | 6.00 | 6.00 | - | 1.00 | 2.00 | - | - | 2.00 | 1.00 | - | - | - | - |
| ...............................................................................TGATGACTGAACGAACTCCTGGGC................................................................................................................................................... | 24 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................TGTGCTGGGAGTTCTCAAAGGCGGGT.................................................................................................... | 26 | 1 | 3.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................TGACTGAACGAACTCCTGGGCATAGACC............................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................................TACAAACCCTGACCTAGTCCTGCGGCC....................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGACTGAACGAACTCCTGGGCATAGAC............................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CTGTGCTGGGAGTTCTCAAAGGCGGGT.................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................ACTGAACGAACTCCTGGGCATAGACCG........................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................TGATGACTGAACGAACTCCTGGtca.................................................................................................................................................. | 25 | tca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................CGAACTCCTGGGCATAGACCGCAG........................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................TGAACGAACTCCTGGGCATAGACCGCAGC....................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TACAAACCCTGACCTAGTCCTGCGGCCT...................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............GAGAGAGATCTAGAGCCCTGGAAA...................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| GTTCCTGGACCGGAGAGAGATCTAGAGCCCTGGAAAGTGGCAGTGAGTGACAGGCTGGTCCTCCATGGGCTTTGGTGCCTGATGACTGAACGAACTCCTGGGCATAGACCGCAGCTGCCTTGGCTGTGCTGGGAGTTCTCAAAGGCGGGTGAGTGGCCCTCTACAGGGTACAAACCCTGACCTAGTCCTGCGGCCTGCAGCCCCGAGGCAGAGCGAGCTCGGCCCCGGCAGGCCCGGCCTGCAGCCCCGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................CGAGCTCGGCCCCGGggc...................... | 18 | ggc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................GCTGCCTTGGCTGTGCTGGGAGTTCTCA............................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |