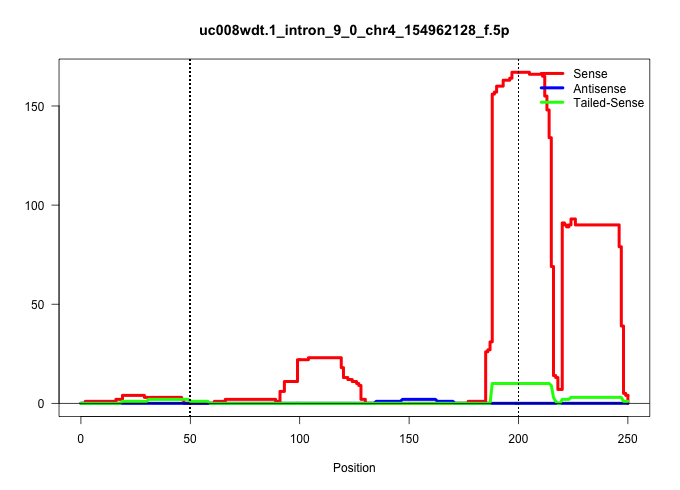
| Gene: Nadk | ID: uc008wdt.1_intron_9_0_chr4_154962128_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
| ACTCGCTGTCATTCCGGCCTATCGTGGTCCCTGCAGGGGTCGAGCTGAAGGTCAGAGCAGTCCCTCTCTGTCTCCTCTGATCTGTGAGTCTTTTGACCCTAGGCTAGCAGAACACAGTTGTTACTACCTTAGCACCTGCCATCTCCAAAGTCTCCCTCCCAGAACCCTCACATGTGAGCCCCACCTGTTAGTGTACCTAGAACCTAAAGCAGACGGGCTCTGCATCTGTCCATGGTCTCTGACGGCACAG ..................................................................................................((((((((((((....(((.(((((......))))).)))............................((((....))))......)))))))....))))).................................................. ..............................................................................................95................................................................................................................209....................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT4() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................TAGTGTACCTAGAACCTAAAGCAGACG................................... | 27 | 1 | 63.00 | 63.00 | 23.00 | 17.00 | 8.00 | 2.00 | 6.00 | 1.00 | 2.00 | - | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TAGTGTACCTAGAACCTAAAGCAGACGG.................................. | 28 | 1 | 51.00 | 51.00 | 16.00 | 13.00 | 6.00 | 4.00 | 2.00 | 4.00 | 1.00 | 1.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGCATCTGTCCATGGTCTCTGACGGCA... | 27 | 1 | 40.00 | 40.00 | 11.00 | 14.00 | 6.00 | 7.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGCATCTGTCCATGGTCTCTGACGGCAC.. | 28 | 1 | 34.00 | 34.00 | 10.00 | 5.00 | 8.00 | 8.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................................................................................................TGCATCTGTCCATGGTCTCTGACGGC.... | 26 | 1 | 11.00 | 11.00 | 1.00 | 7.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TGTTAGTGTACCTAGAACCTAAAGCAG...................................... | 27 | 1 | 9.00 | 9.00 | - | 1.00 | 2.00 | 1.00 | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TGTTAGTGTACCTAGAACCTAAAGCAGAC.................................... | 29 | 1 | 9.00 | 9.00 | 2.00 | 3.00 | - | - | 1.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TAGGCTAGCAGAACACAGTTGTTACTACC.......................................................................................................................... | 29 | 1 | 7.00 | 7.00 | 1.00 | 4.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TAGTGTACCTAGAACCTAAAGCAGACGt.................................. | 28 | t | 6.00 | 63.00 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TAGTGTACCTAGAACCTAAAGCAGACGGGC................................ | 30 | 1 | 5.00 | 5.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................................................TGTTAGTGTACCTAGAACCTAAAGCAGA..................................... | 28 | 1 | 5.00 | 5.00 | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TAGTGTACCTAGAACCTAAAGCAGAC.................................... | 26 | 1 | 5.00 | 5.00 | - | 3.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TCTGTCCATGGTCTCTGACGGCACAG | 26 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TTTGACCCTAGGCTAGCAGAACACAGTTG.................................................................................................................................. | 29 | 1 | 4.00 | 4.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGACCCTAGGCTAGCAGAACACAGTT................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TTAGTGTACCTAGAACCTAAAGCAGACG................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TAGTGTACCTAGAACCTAAAGCAGACGGt................................. | 29 | t | 2.00 | 51.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGCATCTGTCCATGGTCTCTGACGGCAt.. | 28 | t | 2.00 | 40.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TGTTAGTGTACCTAGAACCTAAAGCAGACG................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TTTGACCCTAGGCTAGCAGAACACAGTT................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TAGTGTACCTAGAACCTAAAGCAGA..................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TTAGTGTACCTAGAACCTAAAGCAGACGG.................................. | 29 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................GTGTACCTAGAACCTAAAGCAGACGGGC................................ | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGAACCTAAAGCAGACGGGCTCTGCATC........................ | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TATCGTGGTCCCTGCAGGGGTCGAGCTG........................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TAGGCTAGCAGAACACAGTTGTTACTAC........................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TACCTAGAACCTAAAGCAGACGGGCTCT............................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TAGCAGAACACAGTTGTTACTACCTT........................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................CTAGAACCTAAAGCAGACGGGCTCTGCATC........................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TCTGTCCATGGTCTCTGACGGCACAGt | 27 | t | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................CCTAGAACCTAAAGCAGACGGGCT............................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................GTTAGTGTACCTAGAACCTAAAGCA....................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GCCTATCGTGGTCCCTGCAGGGGTCGAGCT............................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TGCAGGGGTCGAGCTGAAGGTCAGAaaa............................................................................................................................................................................................... | 28 | aaa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TACCTAGAACCTAAAGCAGACGGGCTCTGC........................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................................................TAGTGTACCTAGAACCTAAAGCAGAag................................... | 27 | ag | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TATCGTGGTCCCTGCAGGGGTCGAGCTGtaa........................................................................................................................................................................................................ | 31 | taa | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGACCCTAGGCTAGCAGAACACAGTTG.................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................................................................................TAGAACCTAAAGCAGACGGGCTCTGCA.......................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GCCCCACCTGTTAGTGTACCTAGAACCT............................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................................GTGTACCTAGAACCTAAAGCAGACGG.................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................TAGGCTAGCAGAACACAGTTGTTACTACCTT........................................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TAGTGTACCTAGAACCTAAAGCAGACGGGt................................ | 30 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TACCTAGAACCTAAAGCAGACGGGCTC.............................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TAGTGTACCTAGAACCTAAAGCAG...................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGACCCTAGGCTAGCAGAACACAGTTGTT................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TAGGCTAGCAGAACACAGTTGTTAC.............................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..TCGCTGTCATTCCGGCCTATCGTGGTC............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................ATCTGTCCATGGTCTCTGACGGCACA. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TACCTAGAACCTAAAGCAGACGGGCTCTG............................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TAGGCTAGCAGAACACAGTTGTTACTA............................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................AGTGTACCTAGAACCTAAAGCAGACGG.................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TAGTGTACCTAGAACCTAAAGCAGACGGG................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TCTGTCTCCTCTGATCTGTGAGTCT............................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CCCTCTCTGTCTCCTCTGATCTGTGAGT................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ACTCGCTGTCATTCCGGCCTATCGTGGTCCCTGCAGGGGTCGAGCTGAAGGTCAGAGCAGTCCCTCTCTGTCTCCTCTGATCTGTGAGTCTTTTGACCCTAGGCTAGCAGAACACAGTTGTTACTACCTTAGCACCTGCCATCTCCAAAGTCTCCCTCCCAGAACCCTCACATGTGAGCCCCACCTGTTAGTGTACCTAGAACCTAAAGCAGACGGGCTCTGCATCTGTCCATGGTCTCTGACGGCACAG ..................................................................................................((((((((((((....(((.(((((......))))).)))............................((((....))))......)))))))....))))).................................................. ..............................................................................................95................................................................................................................209....................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT4() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................AAGTCTCCCTCCCAGAACCCTCAC............................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CTGCCATCTCCAAAGTCTCCCTCCCAGA....................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................CCCAGAACCCTCACATgca............................................................................. | 19 | gca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |