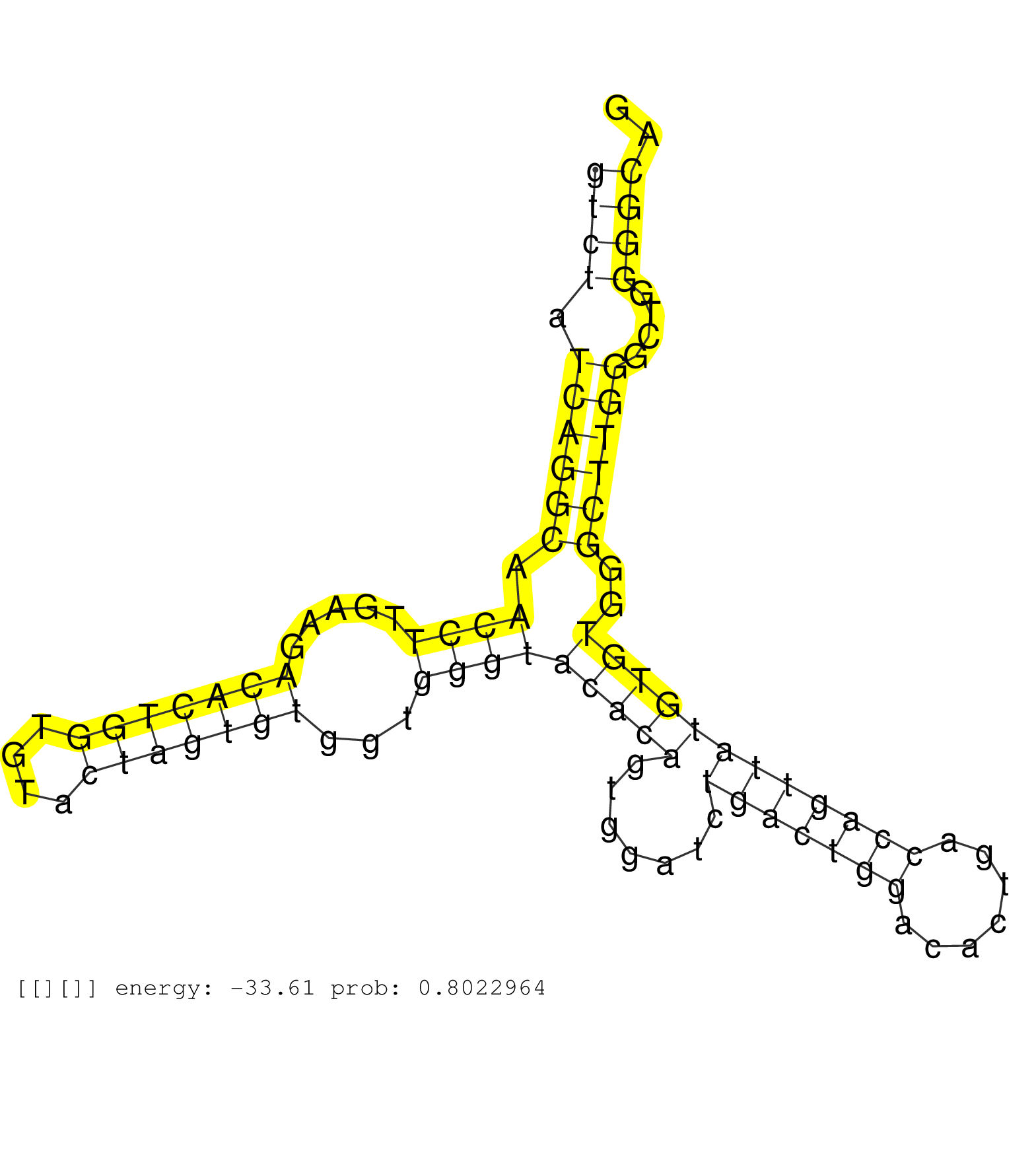

| Gene: Pank4 | ID: uc008wcl.1_intron_6_0_chr4_154345664_f.3p | SPECIES: mm9 |
 |
 |
|
(8) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(30) TESTES |
| AGTAGCAAGCTCAAGGTGGCCTTGTGGAGGCTGTCTTCCTGTCAGGTTCCACAGTGGACAGGCTAGCCAGTGGCATGCAGGTACTCTCTGTTGGCAGGGTCTATCAGGCAACCTTGAAGACACTGGTGTACTAGTGTGGTGGGTACACAGTGGATCTGACTGGACACTGACCAGTTATGTGTGGGCTTGGGCTGGGGCAGGGTGAAGTCCAGGCACTCTTCCTGAGACATGAAGGCTACCTGGGAGCCAT ..................................................................................................((((.((((((.((((.....(((((((....)))))))...))))(((((.......(((((((.......))))))))))))..))))))....)))).................................................... ..................................................................................................99...................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................GTGTGGGCTTGGGCTGGGGCAG.................................................. | 22 | 1 | 550.00 | 550.00 | 162.00 | 127.00 | 78.00 | 37.00 | 42.00 | 16.00 | 15.00 | 23.00 | 20.00 | 7.00 | 3.00 | 2.00 | 1.00 | 4.00 | 2.00 | 2.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 |
| .................................................................................................................................................................................TGTGTGGGCTTGGGCTGGGGCAG.................................................. | 23 | 1 | 165.00 | 165.00 | 51.00 | 45.00 | 16.00 | 14.00 | 2.00 | 13.00 | 11.00 | 3.00 | 2.00 | - | 1.00 | 3.00 | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................................................GTGTGGGCTTGGGCTGGGGCAGt................................................. | 23 | t | 21.00 | 550.00 | 7.00 | - | - | - | 1.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................GTGGGCTTGGGCTGGGGCAG.................................................. | 20 | 1 | 20.00 | 20.00 | 10.00 | - | 6.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TATGTGTGGGCTTGGGCTGGGGCAG.................................................. | 25 | 1 | 8.00 | 8.00 | 2.00 | - | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GTGTGGGCTTGGGCTGGGGCA................................................... | 21 | 1 | 5.00 | 5.00 | - | - | - | - | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GTGTGGGCTTGGGCTGGGGCAa.................................................. | 22 | a | 3.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGTGGGCTTGGGCTGGGGCAG.................................................. | 21 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTGTGGGCTTGGGCTGGGGC.................................................... | 21 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TTATGTGTGGGCTTGGGCTGGGGCAGag................................................ | 28 | ag | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................ATGTGTGGGCTTGGGCTGGGGCAG.................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TATGTGTGGGCTTGGGCTGGGGC.................................................... | 23 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................GTGGGCTTGGGCTGGGGCAGt................................................. | 21 | t | 2.00 | 20.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TATGTGTGGGCTTGGGCTGGGG..................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GTGTGGGCTTGGGCTGGGGC.................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TCAGGCAACCTTGAAGACACTGGTGT......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTGTGGGCTTGGGCTGGGtcgg.................................................. | 23 | tcgg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GTGTGGGCTTGGGCTGGGGCAGcac............................................... | 25 | cac | 1.00 | 550.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTGTGGGCTTGGGCTGGGGaag.................................................. | 23 | aag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GTGTGGGCTTGGGCTGGGacag.................................................. | 22 | acag | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTGTGGGCTTGGGCTGGGGCAGa................................................. | 24 | a | 1.00 | 165.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GTGTGGGCTTGGGCTGGGG..................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................GTGGATCTGACTGGACACTGACCAGTTA......................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CTGAGACATGAAGGCatt........... | 18 | att | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................TGTGTGGGCTTGGGCTGGGGCA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TCCTGAGACATGAAGGCTACCTGGGA..... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................GTGTGGGCTTGGGCTGGGGCgcg................................................. | 23 | gcg | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................GCTTGGGCTGGGGCAG.................................................. | 16 | 10 | 0.30 | 0.30 | - | - | - | - | - | - | 0.30 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGTAGCAAGCTCAAGGTGGCCTTGTGGAGGCTGTCTTCCTGTCAGGTTCCACAGTGGACAGGCTAGCCAGTGGCATGCAGGTACTCTCTGTTGGCAGGGTCTATCAGGCAACCTTGAAGACACTGGTGTACTAGTGTGGTGGGTACACAGTGGATCTGACTGGACACTGACCAGTTATGTGTGGGCTTGGGCTGGGGCAGGGTGAAGTCCAGGCACTCTTCCTGAGACATGAAGGCTACCTGGGAGCCAT ..................................................................................................((((.((((((.((((.....(((((((....)))))))...))))(((((.......(((((((.......))))))))))))..))))))....)))).................................................... ..................................................................................................99...................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|