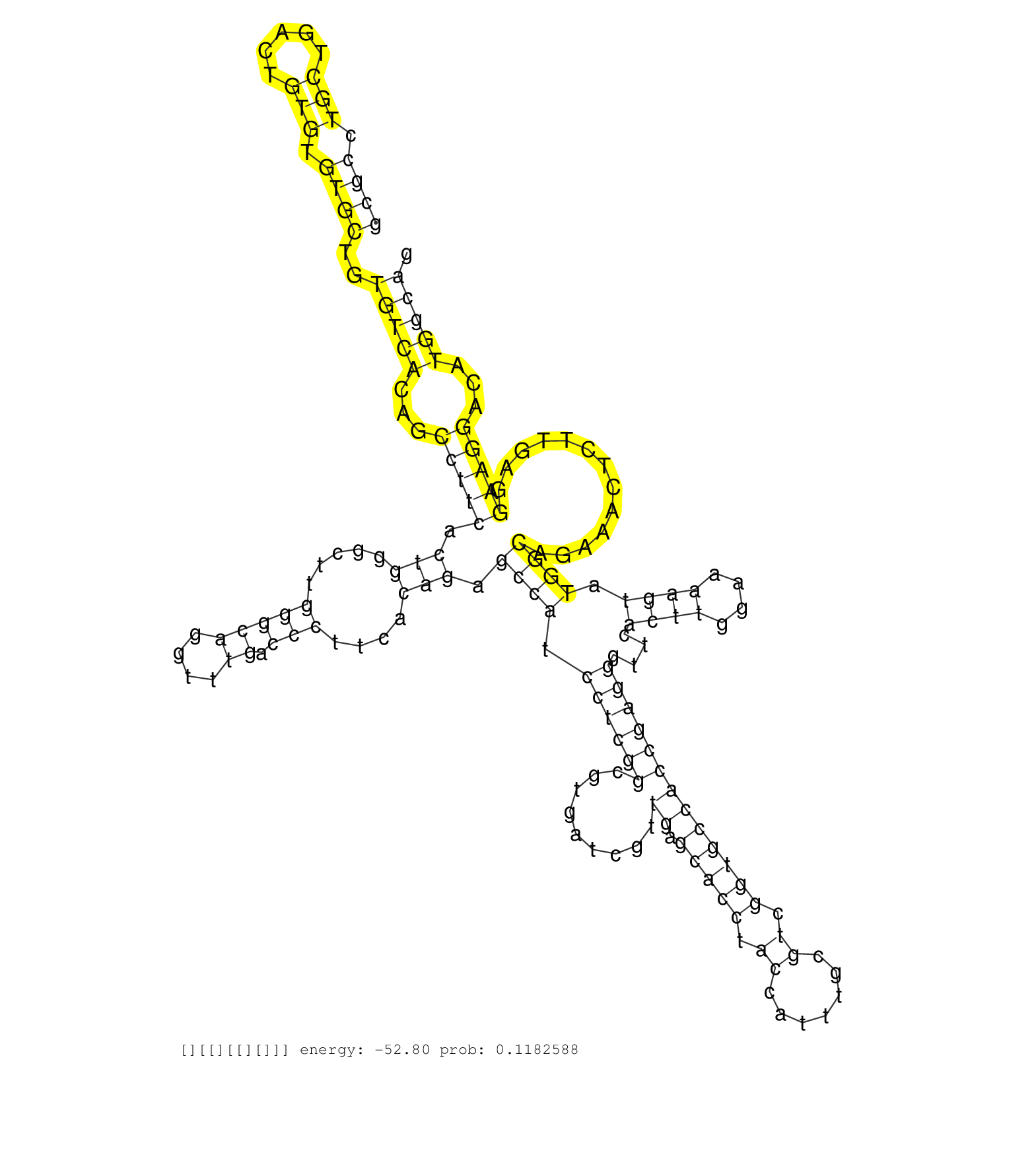

| Gene: Pank4 | ID: uc008wcl.1_intron_3_0_chr4_154344287_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(3) PIWI.mut |
(18) TESTES |
| TCTTCCCCTACCTCCTAGTCAACATTGGCTCTGGCGTCTCCATCGTGAAGGTAAGAGTCCACAAAATGACAGCTGAATGTCTCAGCGCCTGCTGACTGTGTGTGCTGTGTCACAGCCTTCACTGGGCTTGGGCAGGTTTGACCCTTCACAGAGCCATCCTCGGCGTGATCGTTGAGCACCTACCATTTGCGTCGGTGCCACCGAGGGTTCACTTGGAAAAGTATGGCAGAAACTCTTGAGGAAGGACATGGCAGCCCTGACCCCAGCATCCTAGCAGCCTGTGTGGCCTGCAGCCCAGTGCATGCCTTTTCTTCCTCAGGTGGAGACAGAGGACCGGTTTGAGTGGATTGGTGGAAGCTCCATTGGAGG ....................................................................................((((.(((.....))).))))..(((((...(((((.(((.....(((((....)).)))....))).((((.((((((.........((.(((((.((.......)).)))))))))))))....((((....)))).)))).............)))))...))))).................................................................................................................... ....................................................................................85.......................................................................................................................................................................254................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................TGGCAGAAACTCTTGAGGAAGGACATG....................................................................................................................... | 27 | 1 | 8.00 | 8.00 | - | 3.00 | - | - | - | 1.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGCAGAAACTCTTGAGGAAGGACAT........................................................................................................................ | 26 | 1 | 5.00 | 5.00 | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGCAGAAACTCTTGAGGAAGGACATGGC..................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................................................................TGGCAGAAACTCTTGAGGAAGGACA......................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGCTGACTGTGTGTGCTGTGTCACAGC............................................................................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGCAGAAACTCTTGAGGAAGGACATGG...................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................TCGTGAAGGTAAGAGTCCACAAAATGACAGC........................................................................................................................................................................................................................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................GTATGGCAGAAACTCTTGAGGAAGGAC.......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................................................................................................................................GCAGAAACTCTTGAGGAAGGACATGG...................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................AACTCTTGAGGAAGGACATGGCAGCCC................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................ATCGTTGAGCACCTACCATTTGCGTCG............................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................TGGATTGGTGGAAGCTCCATTGGA.. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................................................................GAAACTCTTGAGGAAGGACATGGCA.................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGACTGTGTGTGCTGTGTCACAGCCTT.......................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGCAGAAACTCTTGAGGAAGGAC.......................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAGTCCACAAAATGACAGCTGAAT................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................GGACCGGTTTGAGTGGATTGGTGGAAt............ | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................................................................................AGTGGATTGGTGGAAGCTCCATTGGA.. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................TGCTGACTGTGTGTGCTGTGTCACAGCC............................................................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................TGCATGCCTTTTCTTCCTCAGa................................................. | 22 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGCAGAAACTCTTGAGGAAGGACATa....................................................................................................................... | 27 | a | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................................................................................AGTGGATTGGTGGtgta........... | 17 | tgta | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................TTGAGGAAGGACATGGCAGCCCT............................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGCTGACTGTGTGTGC........................................................................................................................................................................................................................................................................ | 16 | 10 | 0.10 | 0.10 | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCTTCCCCTACCTCCTAGTCAACATTGGCTCTGGCGTCTCCATCGTGAAGGTAAGAGTCCACAAAATGACAGCTGAATGTCTCAGCGCCTGCTGACTGTGTGTGCTGTGTCACAGCCTTCACTGGGCTTGGGCAGGTTTGACCCTTCACAGAGCCATCCTCGGCGTGATCGTTGAGCACCTACCATTTGCGTCGGTGCCACCGAGGGTTCACTTGGAAAAGTATGGCAGAAACTCTTGAGGAAGGACATGGCAGCCCTGACCCCAGCATCCTAGCAGCCTGTGTGGCCTGCAGCCCAGTGCATGCCTTTTCTTCCTCAGGTGGAGACAGAGGACCGGTTTGAGTGGATTGGTGGAAGCTCCATTGGAGG ....................................................................................((((.(((.....))).))))..(((((...(((((.(((.....(((((....)).)))....))).((((.((((((.........((.(((((.((.......)).)))))))))))))....((((....)))).)))).............)))))...))))).................................................................................................................... ....................................................................................85.......................................................................................................................................................................254................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................GGCGTCTCCATCGTGAAGGTAAGAGTCCA.................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |