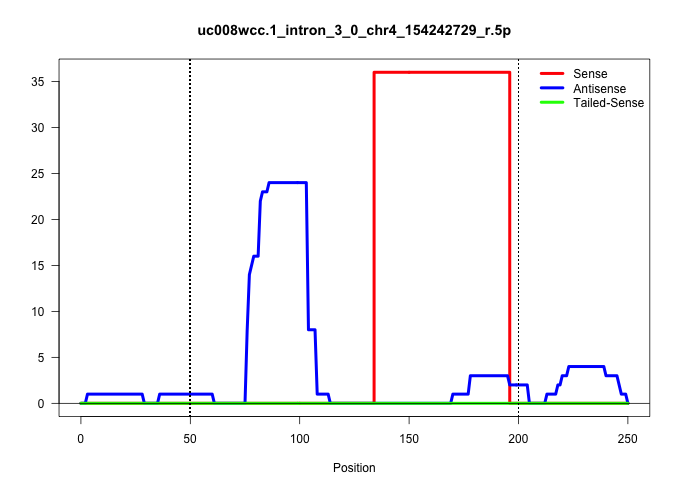
| Gene: AK030206 | ID: uc008wcc.1_intron_3_0_chr4_154242729_r.5p | SPECIES: mm9 |
 |
 |
|
(5) PIWI.ip |
(10) TESTES |
| GTCTCTTGACTCTAGCCCAAAGCGACCCAGGGTCTGCTAGCCTGACCCTGGTAGGTTTCTATGGGCTGGTATGAGTGGCTCCTCCCTGGGTGTTCTTCCTTGCAGTCACTGGGTGGGGTAAGGGGGTTGGTTATTGGGGGCTTACCCATGTAAGGGAGAACCTGCCCATCGTGATGGACGGGCACAGCAGTTGGAAGGGTCCTCACAGGAGATGATGGTGAGAGGCTGCACTGCTCGTCAGTTCTGTCTC ....................................................................................................(((.(((((((((..((.((...((((.((((......)))).))))...))........))..))))..)))))......))).................................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................TGGGGGCTTACCCATGTAAGGGAGAACCTGCCCATCGTGATGGACGGGCACA................................................................ | 52 | 1 | 36.00 | 36.00 | 36.00 | - | - | - | - | - | - | - | - | - |
| GTCTCTTGACTCTAGCCCAAAGCGACCCAGGGTCTGCTAGCCTGACCCTGGTAGGTTTCTATGGGCTGGTATGAGTGGCTCCTCCCTGGGTGTTCTTCCTTGCAGTCACTGGGTGGGGTAAGGGGGTTGGTTATTGGGGGCTTACCCATGTAAGGGAGAACCTGCCCATCGTGATGGACGGGCACAGCAGTTGGAAGGGTCCTCACAGGAGATGATGGTGAGAGGCTGCACTGCTCGTCAGTTCTGTCTC ....................................................................................................(((.(((((((((..((.((...((((.((((......)))).))))...))........))..))))..)))))......))).................................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................GGCTCCTCCCTGGGTGTTCTTCCTTGCA.................................................................................................................................................. | 28 | 1 | 8.00 | 8.00 | - | 3.00 | 2.00 | 2.00 | 1.00 | - | - | - | - | - |
| ..................................................................................TCCCTGGGTGTTCTTCCTTGCAGTCA.............................................................................................................................................. | 26 | 1 | 6.00 | 6.00 | - | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - |
| .............................................................................GCTCCTCCCTGGGTGTTCTTCCTTGCA.................................................................................................................................................. | 27 | 1 | 6.00 | 6.00 | - | 4.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................................CGGGCACAGCAGTTGGAAGGGTCCTCA............................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - |
| .............................................................................GCTCCTCCCTGGGTGTTCTTCCTTGCAGTCA.............................................................................................................................................. | 31 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - |
| ..........................................................................................................................................................................................................................TGAGAGGCTGCACTGCTCGTCAGTTCTG.... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................AGAGGCTGCACTGCTCGTCAGTTCTGT... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................CCCTGGGTGTTCTTCCTTGCAGTCA.............................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GGCTGCACTGCTCGTCAGTTCTGTCTC | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................GATGGTGAGAGGCTGCACTGCTCGTCA.......... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................CTCCTCCCTGGGTGTTCTTCCTTGCA.................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................CTAGCCTGACCCTGGTAGGTTTCTA............................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................................................................................................GTGATGGACGGGCACAGCAGTTGGAA...................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................TGGGTGTTCTTCCTTGCAGTCACTGGGT........................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................TCCTCCCTGGGTGTTCTTCCTTGCA.................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ...TCTTGACTCTAGCCCAAAGCGACCCA............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |