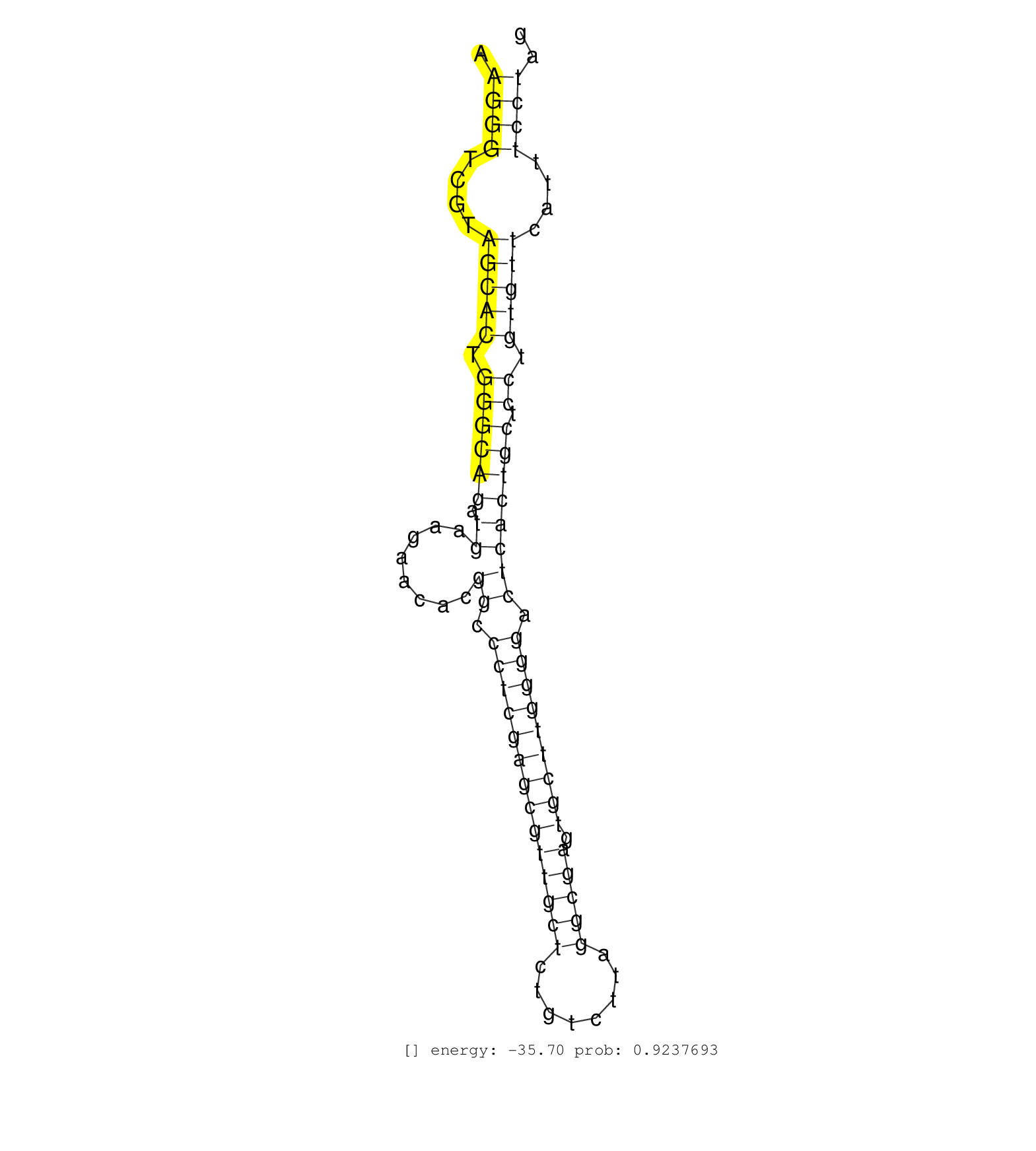
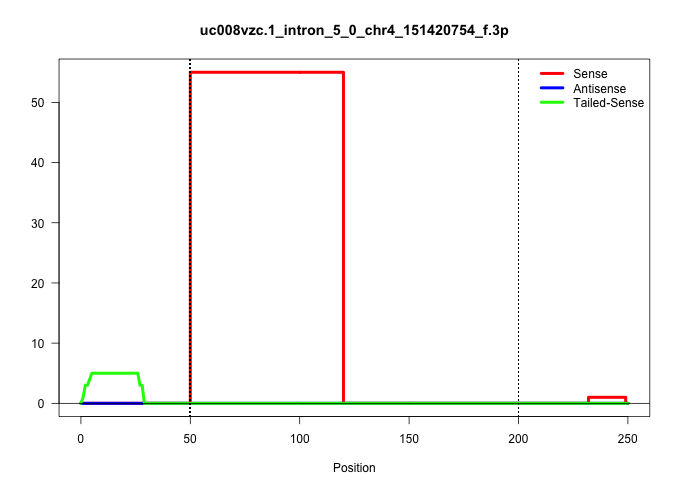
| Gene: Nol9 | ID: uc008vzc.1_intron_5_0_chr4_151420754_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(2) PIWI.ip |
(12) TESTES |
| GTTCCAGGACAGCCAGGGCTACACAGAGAAACCCTGTCTTGAAAAAACAAAAACGAAAAAGTTTTGTGGTTGAGGGTCCCTTCCAAGGTGAACTGTAATAAAGGGTCGTAGCACTGGGCAGATGAAGAACACGGCCCTCGAGCGTTGCTCTGTCTTAGGCGAGTGCTTGGGGACTCACTGCTCCTGTGTTCATTTCCTAGGACCGCCGTACACTCACCAGAGGAAACCACAGAGGATGGTGTACTATGGG .....................................................................................................((((....(((((.((((((.((........((.((((((((((((((........))))).))))))))).)))))))).)).)))))....)))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................AAACGAAAAAGTTTTGTGGTTGAGGGTCCCTTCCAAGGTGAACTGTAATAAA.................................................................................................................................................... | 52 | 1 | 58.00 | 58.00 | 58.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....AGGACAGCCAGGGCTACACAGAGAAACgcag...................................................................................................................................................................................................................... | 31 | gcag | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................CTGGGCAGATGAAGAACA....................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TCGTAGCACTGGGCAGATG.............................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..TCCAGGACAGCCAGGGCTACACcat............................................................................................................................................................................................................................... | 25 | cat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..TCCAGGACAGCCAGGGCTACACAGcgt............................................................................................................................................................................................................................. | 27 | cgt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................AATAAAGGGTCGTAGCACTGGGCAGATGA............................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....AGGACAGCCAGGGCTACACAGcgt............................................................................................................................................................................................................................. | 24 | cgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....CAGGACAGCCAGGGCTACACcgt............................................................................................................................................................................................................................... | 23 | cgt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .TTCCAGGACAGCCAGGGCTACACAtttt............................................................................................................................................................................................................................. | 28 | tttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................................................................................................AGGATGGTGTACTATGG. | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| GTTCCAGGACAGCCAGGGCTACACAGAGAAACCCTGTCTTGAAAAAACAAAAACGAAAAAGTTTTGTGGTTGAGGGTCCCTTCCAAGGTGAACTGTAATAAAGGGTCGTAGCACTGGGCAGATGAAGAACACGGCCCTCGAGCGTTGCTCTGTCTTAGGCGAGTGCTTGGGGACTCACTGCTCCTGTGTTCATTTCCTAGGACCGCCGTACACTCACCAGAGGAAACCACAGAGGATGGTGTACTATGGG .....................................................................................................((((....(((((.((((((.((........((.((((((((((((((........))))).))))))))).)))))))).)).)))))....)))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............GGGCTACACAGAGAAACCCTGTCTTGAAac............................................................................................................................................................................................................... | 30 | ac | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...GACAGCCAGGGCTACACAGAGAAAgtga........................................................................................................................................................................................................................... | 28 | gtga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...GACAGCCAGGGCTACACAGAGAAAgtaa........................................................................................................................................................................................................................... | 28 | gtaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |