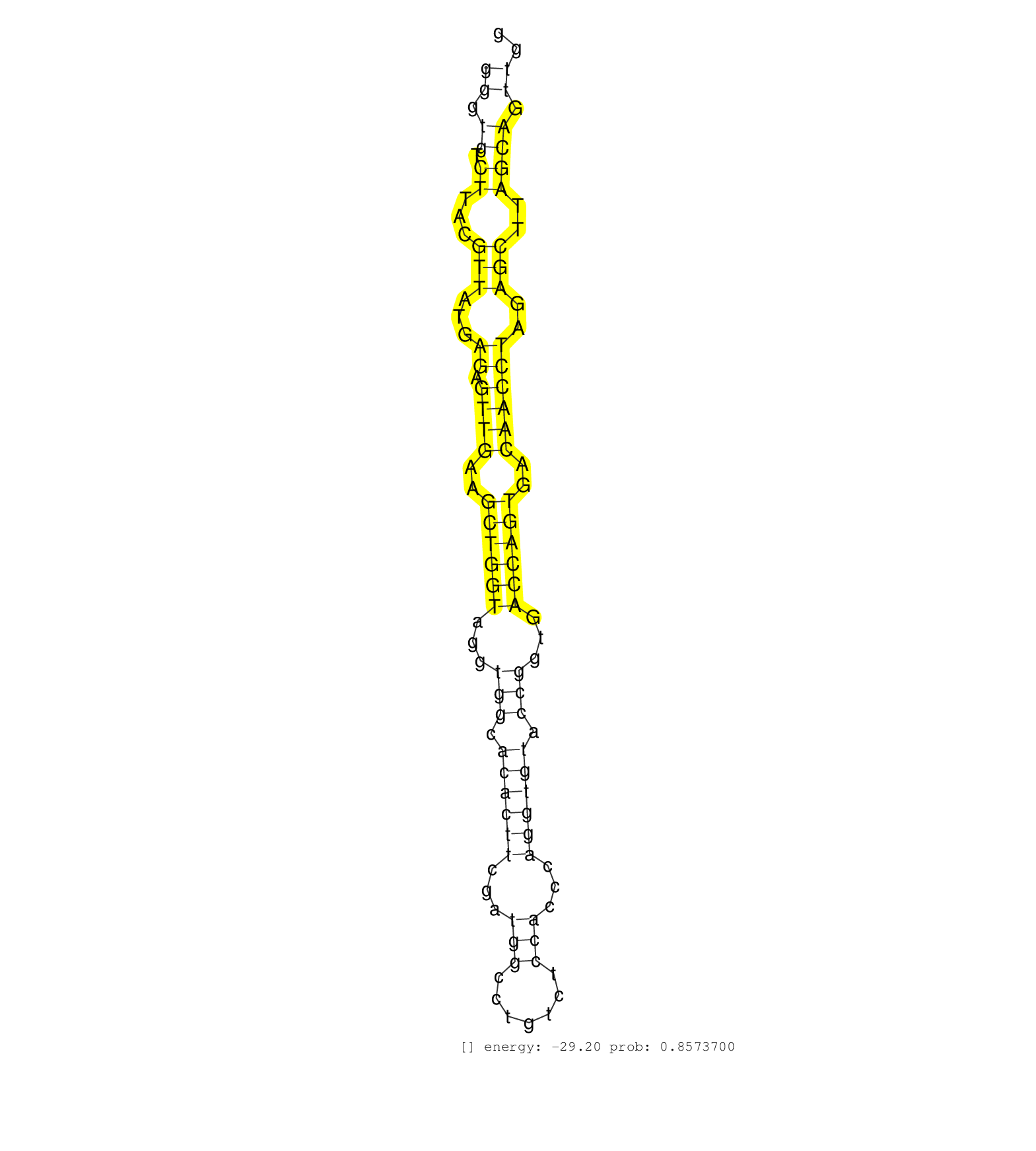

| Gene: Nol9 | ID: uc008vzc.1_intron_1_0_chr4_151415376_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(3) PIWI.mut |
(17) TESTES |
| GGCGATCAGGAGGGAGATCCGGGCCCTGCTCAAGCCTTACACGAAACTAGGTAATTGGCTCCCGCTTCCGATGTCAAGGCTGATGACGAAGGCTTATTATGGGCTGGACGTTGCGCTGATTCCTCATGGGGGGTGTCTTACGTTATGAGAGTTGAAGCTGGTAGGTGGCACACTTCGATGGCCTGTCTCCACCCAGGTGTACCGGTGACCAGTGACAACCTAGAGCTTAGCAGTTGGGACAAGACCCTGCAGGCTCTAGAATTGTCACTGTTGCTGAGGTGTCTTGCTCTTCTTTGGCAGATGACAGAAACTGGGTGGTGCGGTACTTCCCTCCTCTGGGCTCCATTATG ..................................................................................................................................((.((.((...(((...((.((((..((((((...(((.((((((...(((.......)))...)))))).)))...))))))..))))))..)))..)))).))................................................................................................................... ..................................................................................................................................131.......................................................................................................237............................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................TCTTACGTTATGAGAGTTGAAGCTGGT............................................................................................................................................................................................ | 27 | 1 | 4.00 | 4.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................TTACGTTATGAGAGTTGAAGCTGGTA........................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................TATGAGAGTTGAAGCTGGTAGGTGGC..................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TATGAGAGTTGAAGCTGGTAGGTGGCA.................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGAGAGTTGAAGCTGGTAGGTGG...................................................................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TCTTACGTTATGAGAGTTGAAGCTGG............................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................TACGTTATGAGAGTTGAAGCTGGTAt.......................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................GTGTACCGGTGAaatg.......................................................................................................................................... | 16 | aatg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................GATGACAGAAACTGGGTGGTGCGGT.......................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTATGAGAGTTGAAGCTGGTAGGTGGC..................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................GGCTTATTATGGGCTGGACGTTGCGt.......................................................................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................GATGACAGAAACTGGGTGGTGCGGTA......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................................................TACGTTATGAGAGTTGAAGCTGata........................................................................................................................................................................................... | 25 | ata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAATTGGCTCCCGCTTCCGATGTCA.................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................TTCTTTGGCAGATGACAGAAACTGGGT.................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................GACCAGTGACAACCTAGAGCTTAGCAG..................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................................TTATGAGAGTTGAAGCTGGTAG.......................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................TTACGTTATGAGAGTTGAAGCTGGT............................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................AGGCTCTAGAATTGTCACTGTTG............................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................................................TGAGAGTTGAAGCTGGTAGGTGGCAC................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................TGACAGAAACTGGGTGGTGCGGTA......................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGACAACCTAGAGCTTAGCAGTTGGGAC.............................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................GTCTTACGTTATGAGAGTTGAAGCTGG............................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................AACCTAGAGCTTAGCAGTTGGGA............................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TGGACGTTGCGCTGATTCCTCATGGG............................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TCTTACGTTATGAGAGTTGAAGCTGGTA........................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGAGAGTTGAAGCTGGTAGGTGGCA.................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TACGTTATGAGAGTTGAAGCTGGTAGG......................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CTTACGTTATGAGAGTTGAAGCTGGT............................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................GGTGGTGCGGTACTTCCC................... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTATTATGGGCTGGAC................................................................................................................................................................................................................................................. | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |
| GGCGATCAGGAGGGAGATCCGGGCCCTGCTCAAGCCTTACACGAAACTAGGTAATTGGCTCCCGCTTCCGATGTCAAGGCTGATGACGAAGGCTTATTATGGGCTGGACGTTGCGCTGATTCCTCATGGGGGGTGTCTTACGTTATGAGAGTTGAAGCTGGTAGGTGGCACACTTCGATGGCCTGTCTCCACCCAGGTGTACCGGTGACCAGTGACAACCTAGAGCTTAGCAGTTGGGACAAGACCCTGCAGGCTCTAGAATTGTCACTGTTGCTGAGGTGTCTTGCTCTTCTTTGGCAGATGACAGAAACTGGGTGGTGCGGTACTTCCCTCCTCTGGGCTCCATTATG ..................................................................................................................................((.((.((...(((...((.((((..((((((...(((.((((((...(((.......)))...)))))).)))...))))))..))))))..)))..)))).))................................................................................................................... ..................................................................................................................................131.......................................................................................................237............................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................TGACAACCTAGAGCTgtc........................................................................................................................... | 18 | gtc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |