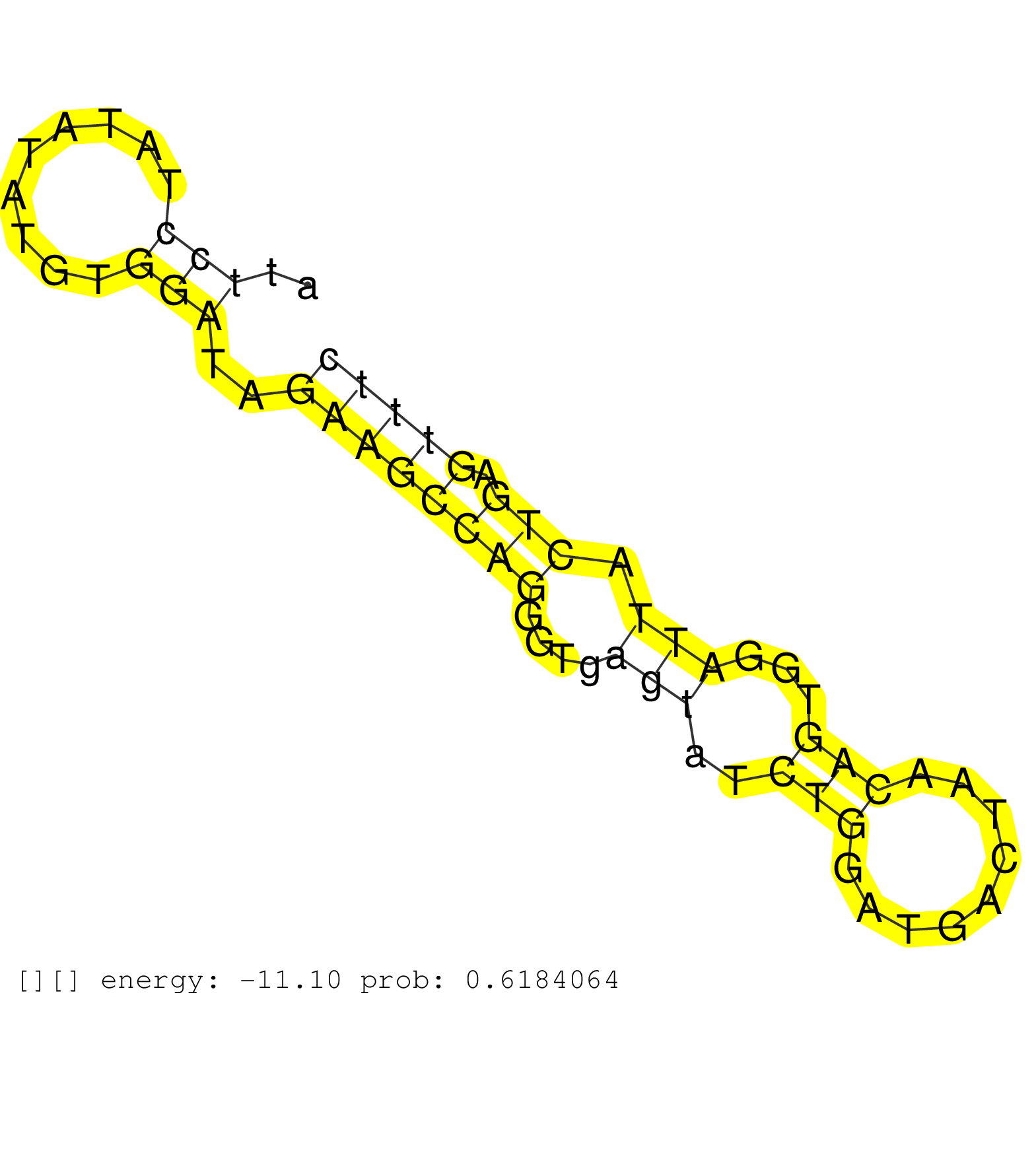

| Gene: Thap3 | ID: uc008vyu.1_intron_2_0_chr4_151359971_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(12) TESTES |
| GACTGGCATGTGGCAAGCTCTGAAGGGCTAGAGGTGGCTCCTGGCATGCGCTTTGGAAGCCTAGTTCCCTCGGGCTCCTTGCCCACGTTTATGGTCCTGGGATTCCTATATATGTGGATAGAAGCCAGGGTGAGTATCTGGATGACTAACAGTGGATTACTGAGTTTCCAAAGCTGTGCCATTGGATGCTGTGCCCACAGGTTCCCCTTCAGCCGCCCGGAGCTGTTGAGGGAGTGGGTGCTCAACATCG .......................................................................................................(((.........)))..((((((((....(((..(((.........)))...))).))).))))).................................................................................. .....................................................................................................102...............................................................168................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................TCTGGATGACTAACAGTGGATTACTGAG...................................................................................... | 28 | 1 | 12.00 | 12.00 | 6.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TCTGGATGACTAACAGTGGATTACTGA....................................................................................... | 27 | 1 | 4.00 | 4.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TATATATGTGGATAGAAGCCAGGGT....................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................TCCAAAGCTGTGCCATTGGATGCTGTG......................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TCTGGATGACTAACAGTGGATTACTGAa...................................................................................... | 28 | a | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TATATATGTGGATAGAAGCCAGGGTG...................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TCCAAAGCTGTGCCATTGGATG.............................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........TGTGGCAAGCTCTGAAGGGCTAGAGGT....................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................ATCTGGATGACTAACAGTGGATTACTGA....................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGCCATTGGATGCTGTGCCCACAGGTTC.............................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................AGCTGTTGAGGGAGTGGGTGCTCAAC.... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................TATATGTGGATAGAAGCCAGGGTGAGT................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................CAGGGTGAGTATCTGGATGA......................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................TCTGGATGACTAACAGTGGATTACTGAGT..................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...TGGCATGTGGCAAGCTCTGAAGGGCTA............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................TAACAGTGGATTACTGAGTTTCCAAAGC............................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................GGGATTCCTATATATGTGGATAGAAGC............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................TATGGTCCTGGGATTCCTATATATGTG...................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| GACTGGCATGTGGCAAGCTCTGAAGGGCTAGAGGTGGCTCCTGGCATGCGCTTTGGAAGCCTAGTTCCCTCGGGCTCCTTGCCCACGTTTATGGTCCTGGGATTCCTATATATGTGGATAGAAGCCAGGGTGAGTATCTGGATGACTAACAGTGGATTACTGAGTTTCCAAAGCTGTGCCATTGGATGCTGTGCCCACAGGTTCCCCTTCAGCCGCCCGGAGCTGTTGAGGGAGTGGGTGCTCAACATCG .......................................................................................................(((.........)))..((((((((....(((..(((.........)))...))).))).))))).................................................................................. .....................................................................................................102...............................................................168................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................GCCCACGTTTATGGTCCTGGGATTCCTATA............................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................CCCTCGGGCTCCTTGCCCACGTTTA............................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................GTCCTGGGATTCCTATATATGTGGATAG................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |