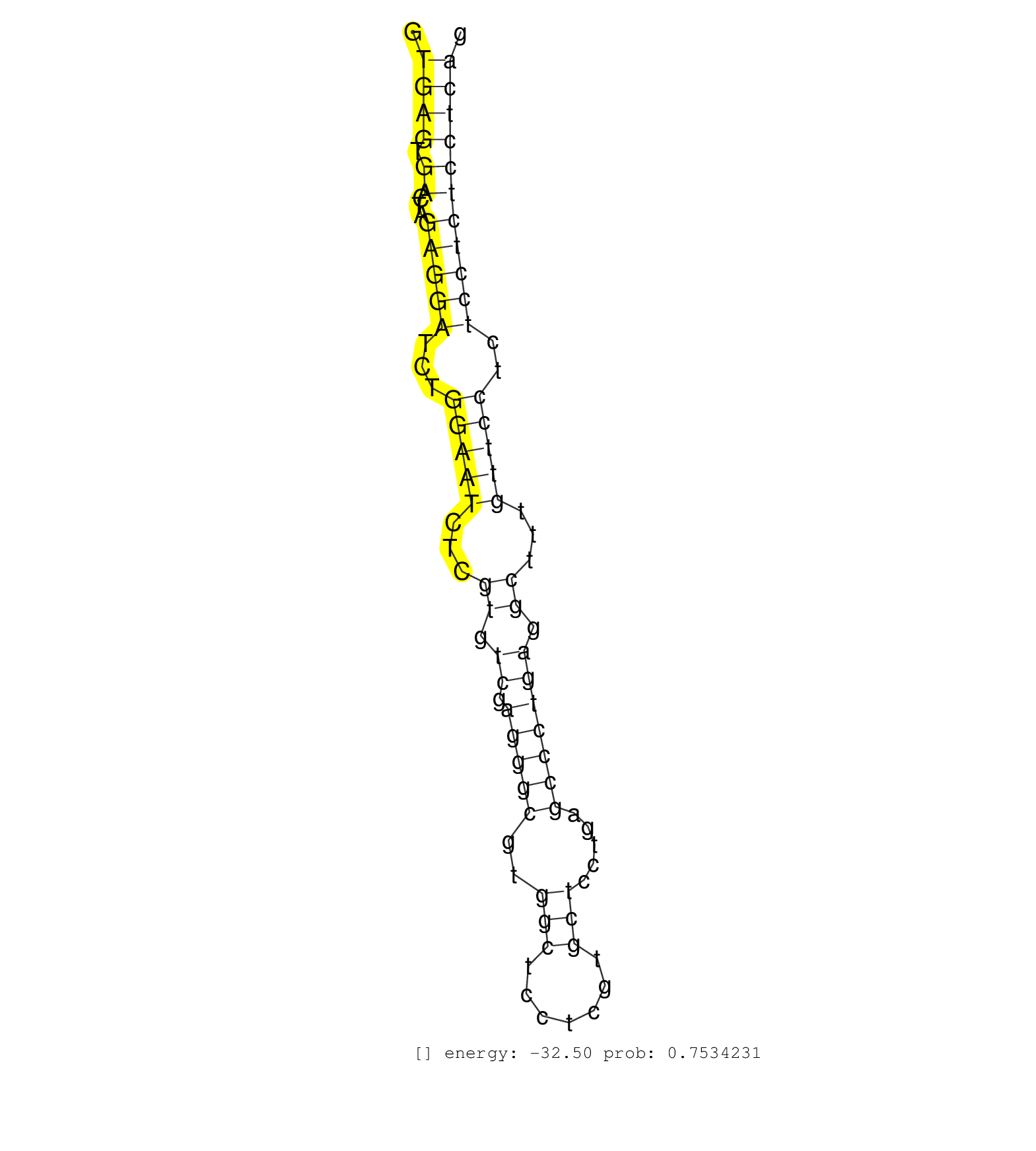

| Gene: Dnajc11 | ID: uc008vys.1_intron_7_0_chr4_151347779_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| CCAGCATCGTCCGGGACACTAAGACCTGCCACTTCACGGTGGCCTTGCAGGTGAGTGACTAGAGGATCTGGAATCTCGTGTCGAGGGCGTGGCTCCTCGTGCTCCTGAGCCCTGAGGCTTTGTTCCTCTCCTCTCCTCAGCTGGGCATCCCTCACTCCTTTGCACTCATCAGTTATCAGCACAAGTTCCA ...................................................((((.((...(((((...(((((...((.((.(((((..(((.......))).....))))))).))...)))))..)))))))))))................................................... ..................................................51.......................................................................................140................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTGACTAGAGGATCTGGAATCTC................................................................................................................. | 27 | 1 | 4.00 | 4.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| ..............................................................AGGATCTGGAATCTCGTGTCGAGGGC...................................................................................................... | 26 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 |
| ..................................................GTGAGTGACTAGAGGATCTGGAATCT.................................................................................................................. | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - |
| ...................................................TGAGTGACTAGAGGATCTGGAATCTC................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ....................................................................TGGAATCTCGTGTCGAGG........................................................................................................ | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGACTAGAGGATCTGG....................................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGACTAGAGGATC.......................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................CTCGTGTCGAGGGCtg.................................................................................................... | 16 | tg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AGGATCTGGAATCTCGTGTCGAGGGCGTG................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGACTAGAGGATCTGGAATCTCGTGT............................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGTGACTAGAGGATCTGGAATCTCGT............................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TAGAGGATCTGGAATCTCGTGTCGAGG........................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCAGCATCGTCCGGGACACTAAGACCTGCCACTTCACGGTGGCCTTGCAGGTGAGTGACTAGAGGATCTGGAATCTCGTGTCGAGGGCGTGGCTCCTCGTGCTCCTGAGCCCTGAGGCTTTGTTCCTCTCCTCTCCTCAGCTGGGCATCCCTCACTCCTTTGCACTCATCAGTTATCAGCACAAGTTCCA ...................................................((((.((...(((((...(((((...((.((.(((((..(((.......))).....))))))).))...)))))..)))))))))))................................................... ..................................................51.......................................................................................140................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................TCAGCACAAGTTCCA | 15 | 18 | 0.11 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |