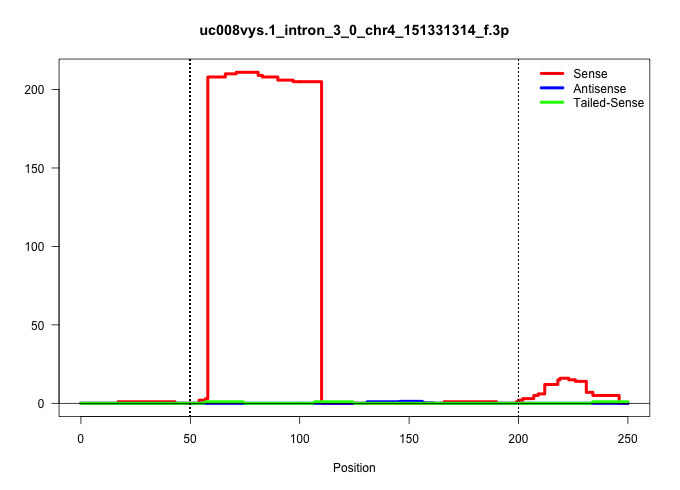
| Gene: Dnajc11 | ID: uc008vys.1_intron_3_0_chr4_151331314_f.3p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(1) TDRD1.ip |
(16) TESTES |
| CCCGTGGAACTGGTGTTTGCTGGCTCTGATAGTCCAAGGCATTCCGCCGCAGTGTTTTATTTGACCTGTGACATTGATGCTGTGTGGGCTTGGGCGCACTGTCCTTCCGTGAAGTCAGATGCCTGAGCACTCTCCCCGCCTCAGCGCTCTCAGTCCACAGACGCTGCACCCAGTGCTGACTGCTTGCTTTTCTGCCCCAGGGAACCATCAGCGTTGGAGTAGATGCGACTGACCTTTTTGATCGTTATGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................ATTTGACCTGTGACATTGATGCTGTGTGGGCTTGGGCGCACTGTCCTTCCGT............................................................................................................................................ | 52 | 1 | 205.00 | 205.00 | 205.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................GTTGGAGTAGATGCGACTG................... | 19 | 1 | 6.00 | 6.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................GTAGATGCGACTGACCTTTTTGATCGTT.... | 28 | 1 | 4.00 | 4.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCAGCGTTGGAGTAGATGCGACTGACC................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..................................................................TGTGACATTGATGCTGTGTGGGCT................................................................................................................................................................ | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................TTTTATTTGACCTGTGACATTGATGCT......................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TAGATGCGACTGACCTTTTTGATCGTT.... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................TATTTGACCTGTGACATTGATGCTGT....................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................AACCATCAGCGTTGGAGTAGA........................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................TTTTTGATCGTTATGcg | 17 | cg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGTGAAGTCAGATGagaa............................................................................................................................. | 18 | agaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................................................................................GGAACCATCAGCGTTGGA................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AGCGTTGGAGTAGATGCGACTG................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................................................GGGAACCATCAGCGTTGGAGTAGATGC........................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................CACCCAGTGCTGACTGCTTGCTTT............................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................TATTTGACCTGTGAtatc............................................................................................................................................................................... | 18 | tatc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................CATTGATGCTGTGTGGGCTTGGGCGC......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TGCTGGCTCTGATAGTCCAAGGCATT............................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| CCCGTGGAACTGGTGTTTGCTGGCTCTGATAGTCCAAGGCATTCCGCCGCAGTGTTTTATTTGACCTGTGACATTGATGCTGTGTGGGCTTGGGCGCACTGTCCTTCCGTGAAGTCAGATGCCTGAGCACTCTCCCCGCCTCAGCGCTCTCAGTCCACAGACGCTGCACCCAGTGCTGACTGCTTGCTTTTCTGCCCCAGGGAACCATCAGCGTTGGAGTAGATGCGACTGACCTTTTTGATCGTTATGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................CTCCCCGCCTCAGCGCTCTCAGTCCA............................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................CTCTCAGTCCACAGAC........................................................................................ | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |