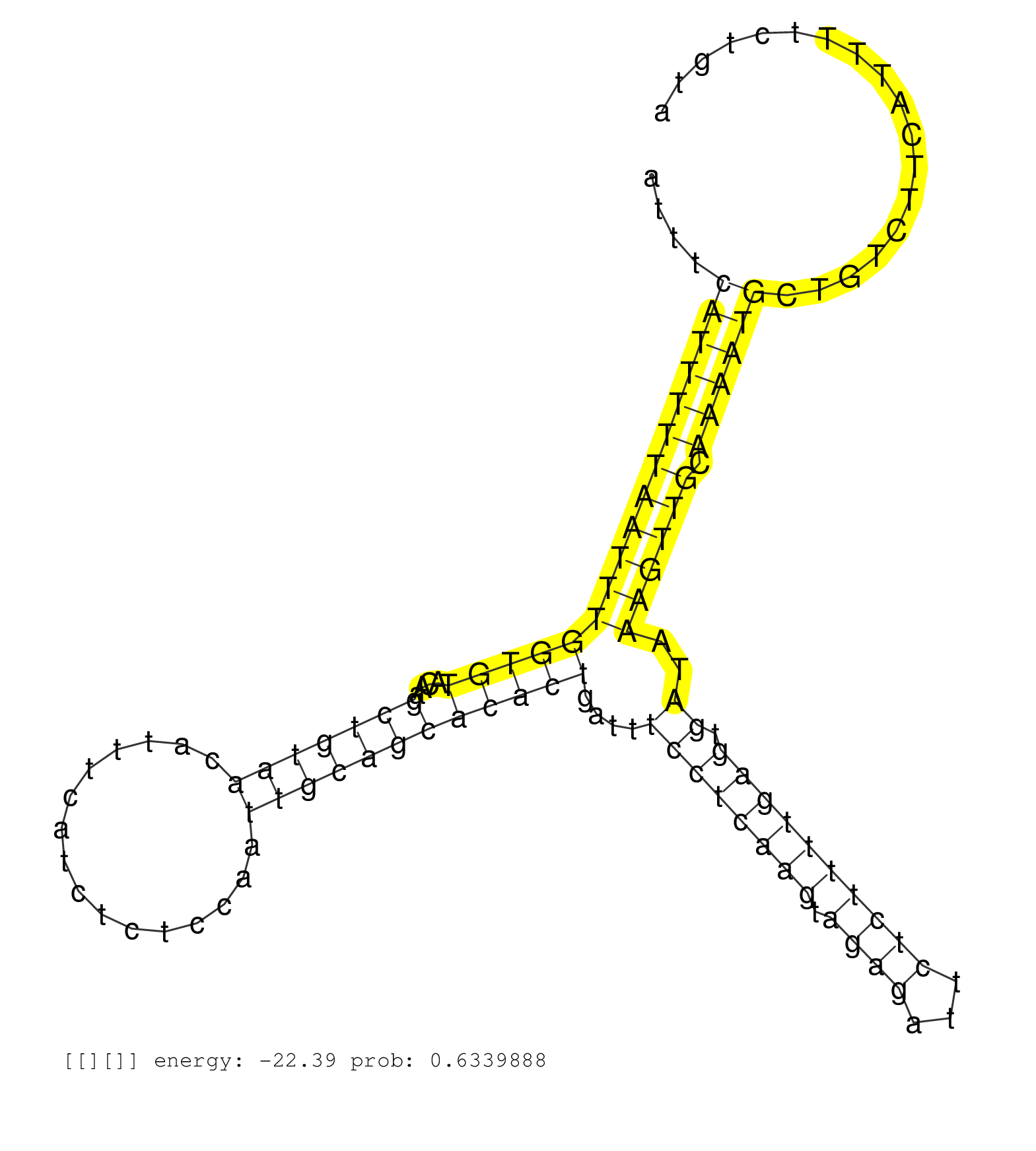

| Gene: Camta1 | ID: uc008vyq.1_intron_1_0_chr4_151164663_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| TTAATAATGTGTGACCCTTCACCTTTGATCCCCTGACCTGCATTACCTTGGTAACCATTTCATTTTTTAATTTAATTTCATTTTTAATTTGGTGTACAAGCTGTAACATTTCATCTCTCCAATTGCAGCACACTGATTTCCTCAAGTAGAGATTCTCTTTTGAGTGATAAAGTTGCAAAATGCTGTCTTCATTTTCTGTATTTAAATTCATTTTGATTTTTAAGAATAAAGTGTAATCTGTGTTTTCATC ..............................................................................(((((((((((((((((....(((((((................))))))))))))....((((((((.((((...)))))))))).))..)))))).)))))).................................................................... ..........................................................................75...........................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................ATTTTTAATTTGGTGgaat........................................................................................................................................................ | 19 | gaat | 4.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - |
| .................TTCACCTTTGATCCCCTGACCTGCATT.............................................................................................................................................................................................................. | 27 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TTGCAGCACACTGATTTCCTCAAGTAGA.................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................ACAAGCTGTAACATTTCATCTCTCCAA................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................ATAAAGTTGCAAAATGCTGTCTTCATTT........................................................ | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................GCAAAATGCTGTCTTCATTTT....................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................TGACCTGCATTACCTTGGTAACCATTTC............................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................CCAATTGCAGCACACTGATTTCCTCAA......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................TGATTTCCTCAAGTAGAGATTCTCTTT.......................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGGTGTACAAGCTGTAACATTTCATCTC..................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TTCACCTTTGATCCCCTGACCTGCAT............................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TCTCCAATTGCAGCACACTGATTTCCTC........................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TTTGGTGTACAAGCTGTAACATTTCA......................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TACAAGCTGTAACATTTCATCTCTCC.................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGGTGTACAAGCTGTAACATTTCATCT...................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................ACAAGCTGTAACATTTCATCTCTCCAATT.............................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................ATTTTTAATTTGGTGggat........................................................................................................................................................ | 19 | ggat | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................TTTGATCCCCTGACCTGCATT.............................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGAGTGATAAAGTTGCAAAATGCTGTC............................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................CACCTTTGATCCCCTGACCTGCATTACC........................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TCACCTTTGATCCCCTGACCTGCATTACC........................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AGTAGAGATTCTCTTTTGA....................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................TCTCTCCAATTGCAGCACACTGATTTC.............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TCTCCAATTGCAGCACACTGATTTCCTCA.......................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....TAATGTGTGACCCTTCACCTTTGATCCCt......................................................................................................................................................................................................................... | 29 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................TTGATCCCCTGACCTGC................................................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................TTCACCTTTGATCCCCTGACCTGCATTACC........................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................TTTGATCCCCTGACCTGCATTACCTTG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTAATAATGTGTGACCCTTCACCTTTGATCCCCTGACCTGCATTACCTTGGTAACCATTTCATTTTTTAATTTAATTTCATTTTTAATTTGGTGTACAAGCTGTAACATTTCATCTCTCCAATTGCAGCACACTGATTTCCTCAAGTAGAGATTCTCTTTTGAGTGATAAAGTTGCAAAATGCTGTCTTCATTTTCTGTATTTAAATTCATTTTGATTTTTAAGAATAAAGTGTAATCTGTGTTTTCATC ..............................................................................(((((((((((((((((....(((((((................))))))))))))....((((((((.((((...)))))))))).))..)))))).)))))).................................................................... ..........................................................................75...........................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) |
|---|