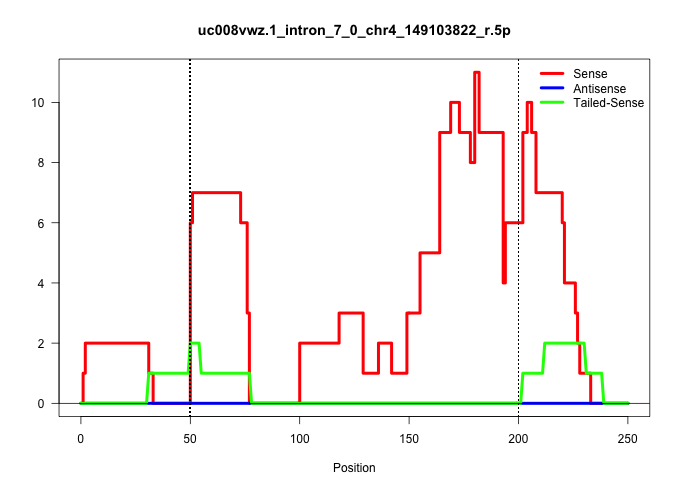
| Gene: D4Ertd429e | ID: uc008vwz.1_intron_7_0_chr4_149103822_r.5p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| ACCAGACCACCAAGATCAAGCAGCTGGCTGCTTTCACACCACGGGAGGAGGTGAGTACACATGGCTGATGGCTGGGTAGAGCCACACTGTCACCTGCCGCTCCAGGGTCCCACCCACATTCCAGGGTGCAGCTTCCTTCACAGCCGAGCTGATCTTGTGACCACTGGGAGAACAACACCATGGAAACCAGAGCCTGCCAGGATAGATGGCTCTGTCCCTGAGGAAAGGGAGGTAGAGGCCCTGGGCTCCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................TGGGAGAACAACACCATGGAAACCAGAGC......................................................... | 29 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACACATGGCTGATGGCTGGG.............................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGGAAACCAGAGCCTGCCAGGATAGATG.......................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGTGACCACTGGGAGAACAACACCATG.................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTGAGTACACATGGCTGATGGCTGGGT............................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TCCAGGGTCCCACCCACATTCCAGGGTGC......................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................TGCCAGGATAGATGGCTCTGTCCCTGA............................. | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TGATCTTGTGACCACTGGGAGAAC............................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TTCACAGCCGAGCTGATCTTGTGACCACT..................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTGAGTACACATGGCTGATGGCT................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TAGATGGCTCTGTCCCTGAGGAAAG....................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..CAGACCACCAAGATCAAGCAGCTGGCTGCTT......................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TGCCAGGATAGATGGCTCTGTCCCTG.............................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGTCCCTGAGGAAAGGGAGGTAGAGGt........... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................TGGAAACCAGAGCCTGCCAGGATAGA............................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TGATCTTGTGACCACTGGGAGAACAACAC........................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTACACATGGCTGATGGCTGGGT............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TTCCAGGGTGCAGCTTCCTTCACA............................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................GAACAACACCATGGAAACCAGAGCC........................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TAGATGGCTCTGTCCCTGAGGAAAGG...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TAGATGGCTCTGTCCCTGAGGAAA........................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACACATGGCTGATGGCTGGGaa............................................................................................................................................................................ | 28 | aa | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TTTCACACCACGGGAGGAGGgccg................................................................................................................................................................................................... | 24 | gccg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TAGATGGCTCTGTCCCTGAGGAAAGGGAa................... | 29 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................GATGGCTCTGTCCCTGAGGAAAGGGAGGT................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GGGAGAACAACACCATGGAAACCAGAGC......................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .CCAGACCACCAAGATCAAGCAGCTGGCTGC........................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ACCAGACCACCAAGATCAAGCAGCTGGCTGCTTTCACACCACGGGAGGAGGTGAGTACACATGGCTGATGGCTGGGTAGAGCCACACTGTCACCTGCCGCTCCAGGGTCCCACCCACATTCCAGGGTGCAGCTTCCTTCACAGCCGAGCTGATCTTGTGACCACTGGGAGAACAACACCATGGAAACCAGAGCCTGCCAGGATAGATGGCTCTGTCCCTGAGGAAAGGGAGGTAGAGGCCCTGGGCTCCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................GAGCCTGCCAGGATgcca............................................... | 18 | gcca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |