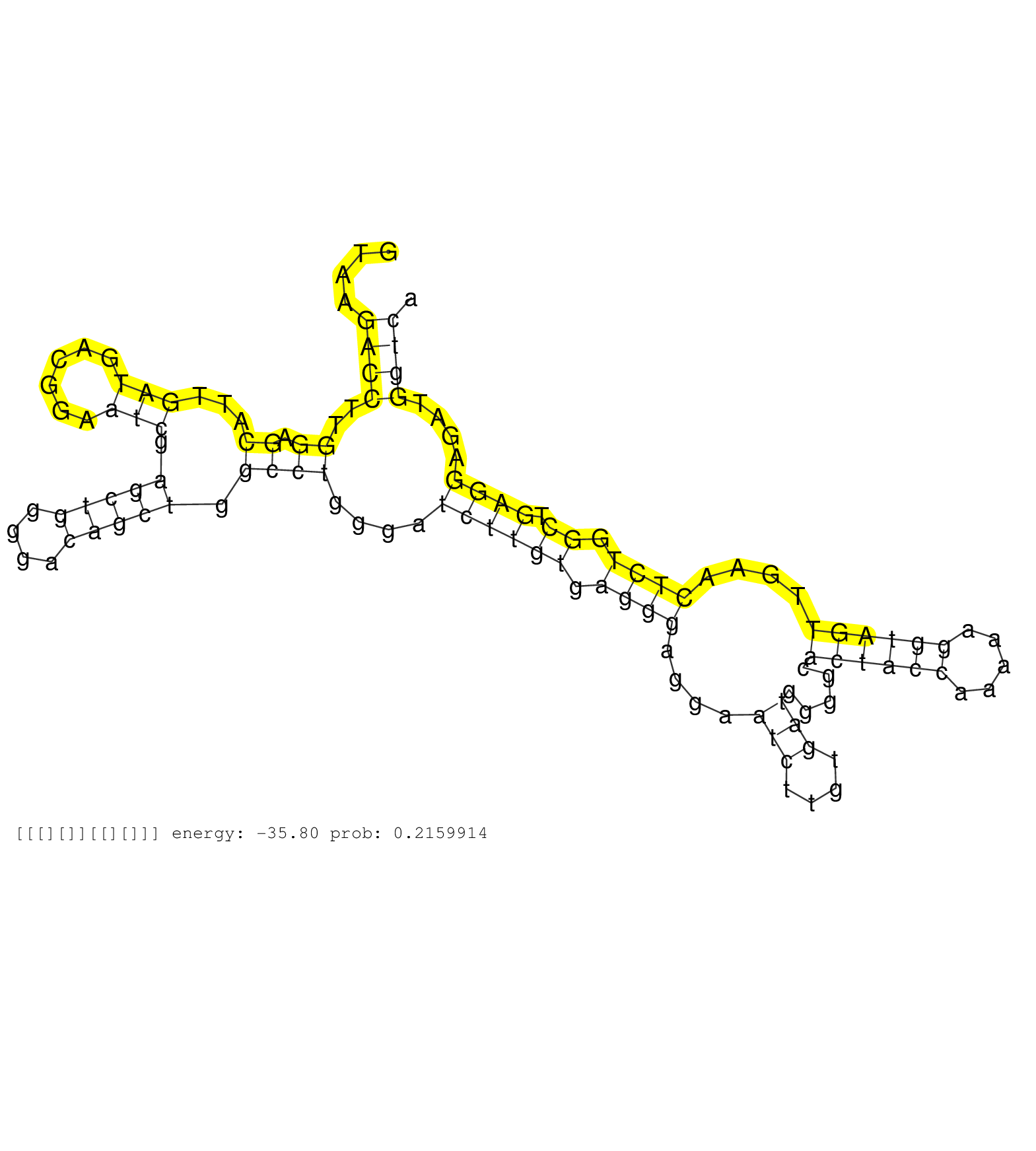
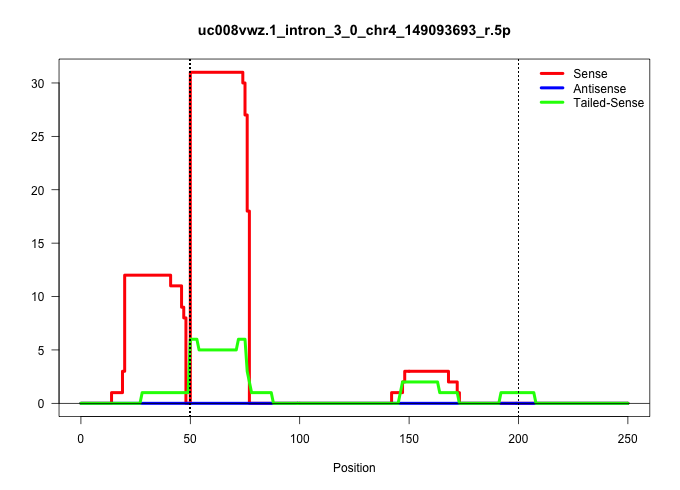
| Gene: D4Ertd429e | ID: uc008vwz.1_intron_3_0_chr4_149093693_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| CCTCAGTCGGGCCCTCTCGCTGGGAACCATACCTCCTCTGACCCGGACTGGTAAGACCTTGGAGCATTGATGACGGAATCGAGCTGGGGACAGCTGGCCTGGGATCTTGTGAGGGAGGAATCTTGTGATGGGGCACTACCAAAAAGGTAGTTGAACTCTGGCTGAGGAGATGGTCAGGGCCAGGCTCAGATCCAGGTGAACCCACTCAGCAGCCACAGTGACAAATCTCTTGTTTGCCTACCCAAACCAT ......................................................((((..((.((...(((......))).(((((....))))).))))....((((((.((((....(((....))).....((((((.....))))))....)))).)).))))....))))........................................................................... ..................................................51...........................................................................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGACCTTGGAGCATTGATGACGGA............................................................................................................................................................................. | 27 | 1 | 21.00 | 21.00 | 7.00 | 2.00 | 3.00 | - | - | 3.00 | 2.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTAAGACCTTGGAGCATTGATGACGG.............................................................................................................................................................................. | 26 | 1 | 12.00 | 12.00 | 2.00 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| ....................TGGGAACCATACCTCCTCTGACCCGGAC.......................................................................................................................................................................................................... | 28 | 1 | 8.00 | 8.00 | - | 4.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTAAGACCTTGGAGCATTGATGACG............................................................................................................................................................................... | 25 | 1 | 5.00 | 5.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTAAGACCTTGGAGCATTGATGACGa.............................................................................................................................................................................. | 26 | a | 3.00 | 5.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CTGGGAACCATACCTCCTCTGACCCGG............................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................AGTTGAACTCTGGCTGAGGAGATG.............................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................ACGGAATCGAGCgctc.................................................................................................................................................................. | 16 | gctc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAAGACCTTGGAGCATTGATGAC................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAGTTGAACTCTGGCTGAGGAGATGG............................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TCTCGCTGGGAACCATACCTCCTCTGA................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAGTTGAACTCTGGCTGAGGAGATGt............................................................................. | 26 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGACCTTGGAGCATTGATGACGaaa............................................................................................................................................................................ | 28 | aaa | 1.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................GTAGTTGAACTCTGGgcc...................................................................................... | 18 | gcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................AAAGGTAGTTGAACTCTGGCTGAGGA.................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CAGGTGAACCCACTgg.......................................... | 16 | gg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................ATACCTCCTCTGACCCGGACTGactc.................................................................................................................................................................................................... | 26 | actc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTAAGACCTTGGAGCATTGATGACGGAA............................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGACCTTGGAGCATTGATGACGaa............................................................................................................................................................................. | 27 | aa | 1.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TGGGAACCATACCTCCTCTGACCCGGA........................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCTCAGTCGGGCCCTCTCGCTGGGAACCATACCTCCTCTGACCCGGACTGGTAAGACCTTGGAGCATTGATGACGGAATCGAGCTGGGGACAGCTGGCCTGGGATCTTGTGAGGGAGGAATCTTGTGATGGGGCACTACCAAAAAGGTAGTTGAACTCTGGCTGAGGAGATGGTCAGGGCCAGGCTCAGATCCAGGTGAACCCACTCAGCAGCCACAGTGACAAATCTCTTGTTTGCCTACCCAAACCAT ......................................................((((..((.((...(((......))).(((((....))))).))))....((((((.((((....(((....))).....((((((.....))))))....)))).)).))))....))))........................................................................... ..................................................51...........................................................................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|