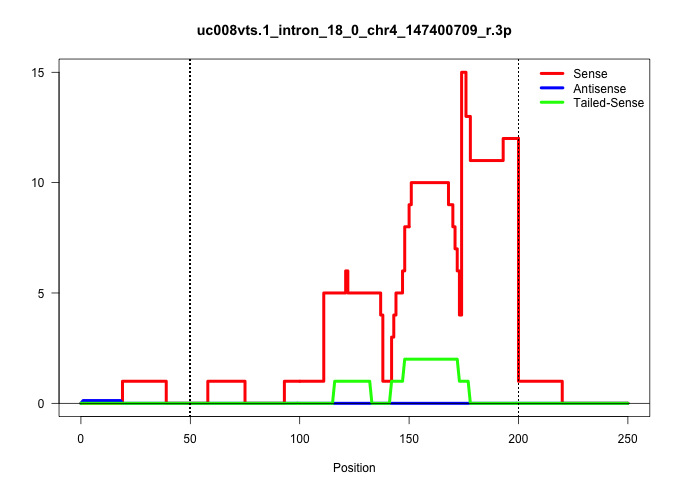
| Gene: Clcn6 | ID: uc008vts.1_intron_18_0_chr4_147400709_r.3p | SPECIES: mm9 |
 |
|
(3) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| TTATTTTCTCTTTTCTTTGGAGAGTCAGTGAGGCTATTCTGGGGCAGGATCTCTCAGCCAGCGTGACTAGCAGAGTTTACTCTCCTGCTGCTGCTGCAGGTGCCTTGAGTGACCACGGAAGTGCAGTGAGGTCAGGTGAAGCTTCCAAGAGACCTGTACTCACCTGGAGATGAAGATGTCCACTCTCTTATCCTCTGCAGGTGGGTCTCTTTGTGGACTTTTCCGTGCGACTCTTCACCCAACTGAAGTT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037903(GSM510439) testes_rep4. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................GATGTCCACTCTCTTATCCTCTGCAG.................................................. | 26 | 1 | 11.00 | 11.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................ACCACGGAAGTGCAGTGAGGTCAGGTG................................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GAGACCTGTACTCACCTGGAGATGAAGA.......................................................................... | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GAGAGTCAGTGAGGCTATTC................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TCTGCAGGTGGGTCTCTTTGTGGACTT.............................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................ACCTGTACTCACCTGGAGATGAAGATG........................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTCCAAGAGACCTGTACTCACCTGGAGATGt............................................................................. | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................CAGCGTGACTAGCAGAG............................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGCAGTGAGGTCAGGTGAAGCTTC......................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................GACCTGTACTCACCTGGAGATGAAGATG........................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................CTGCAGGTGCCTTGAGTGACCACGGAAGT................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................................................GAGACCTGTACTCACCTGGAGATGAAGATt........................................................................ | 30 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................CAAGAGACCTGTACTCACCTGGAGATGA............................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................ACCACGGAAGTGCAGTGAGGTCAGGT................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................CCAAGAGACCTGTACTCACCTGGAGATG.............................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................AGAGACCTGTACTCACCTGGAGATGA............................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTCCAAGAGACCTGTACTCACCTGGAGAT............................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................GGAAGTGCAGTGAtgta..................................................................................................................... | 17 | tgta | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTCCAAGAGACCTGTACTCACCTGGA.................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCCAAGAGACCTGTACTCACCTGGAGA................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| TTATTTTCTCTTTTCTTTGGAGAGTCAGTGAGGCTATTCTGGGGCAGGATCTCTCAGCCAGCGTGACTAGCAGAGTTTACTCTCCTGCTGCTGCTGCAGGTGCCTTGAGTGACCACGGAAGTGCAGTGAGGTCAGGTGAAGCTTCCAAGAGACCTGTACTCACCTGGAGATGAAGATGTCCACTCTCTTATCCTCTGCAGGTGGGTCTCTTTGTGGACTTTTCCGTGCGACTCTTCACCCAACTGAAGTT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037903(GSM510439) testes_rep4. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .TATTTTCTCTTTTCTTTG....................................................................................................................................................................................................................................... | 18 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |