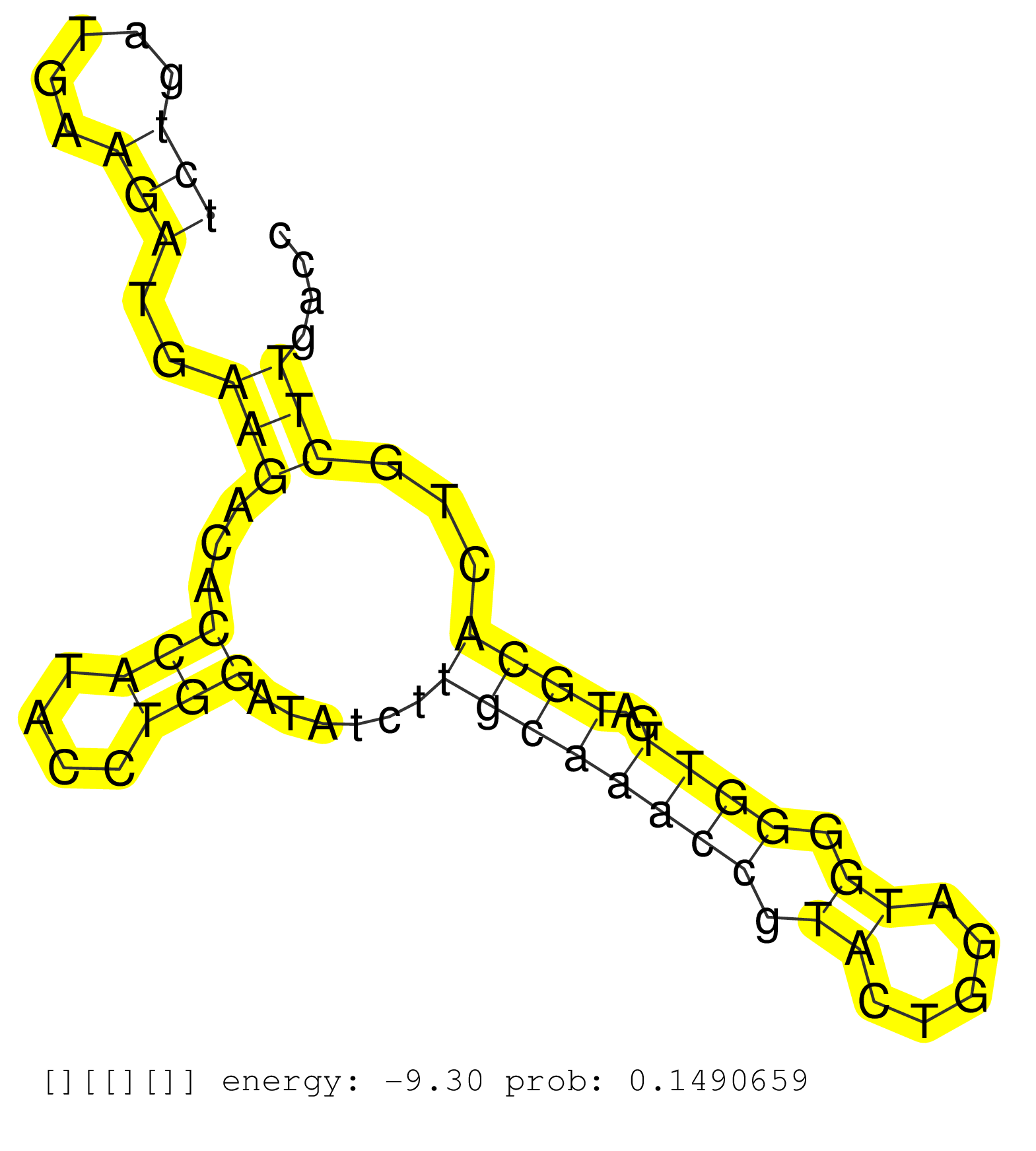

| Gene: Pramef12 | ID: uc008vre.1_intron_1_0_chr4_143985063_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(19) TESTES |
| AAGATGCCATCACTCACAAACGACTTAACATCCTGAGGAAGATGGTGCAGGTCTGGCCTTTTCCCTGTCTCCCGCTGGGTGGTCTGATGAAGATGAAGACACCATACCTGGATATCTTGCAAACCGTACTGGATGGGGTTGATGCACTGCTTGACCAGAATACTCACCCCAGGTGAGGGAGCCCCAGGGACTCTGAAGGGAGGCCTGGTAGTCTAGGGTAGAGAAAGAATAGAGAATGGAAGGAAGGTTA ..................................................................................(((.....)))..(((...(((....)))......((((((((.((.....)).))))..))))...))).................................................................................................. ..................................................................................83.......................................................................156............................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................TGAAGATGAAGACACCATACCTGGATA........................................................................................................................................ | 27 | 1 | 20.00 | 20.00 | 4.00 | 5.00 | 6.00 | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGAAGATGAAGACACCATACCTGGATATC...................................................................................................................................... | 29 | 1 | 18.00 | 18.00 | 2.00 | 5.00 | 5.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGATGAAGATGAAGACACCATACCTGGAT......................................................................................................................................... | 29 | 1 | 14.00 | 14.00 | 9.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TACTGGATGGGGTTGATGCACTGCTT.................................................................................................. | 26 | 1 | 13.00 | 13.00 | 4.00 | 2.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................TGATGCACTGCTTGACCAGAATACTCACC.................................................................................. | 29 | 1 | 11.00 | 11.00 | 5.00 | 3.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGAAGATGAAGACACCATACCTGGAT......................................................................................................................................... | 26 | 1 | 7.00 | 7.00 | 2.00 | - | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGATGAAGATGAAGACACCATACCTGGATA........................................................................................................................................ | 30 | 1 | 4.00 | 4.00 | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGATGCACTGCTTGACCAGAATACTCACCC................................................................................. | 30 | 1 | 4.00 | 4.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................CAAACGACTTAACATCCTGAGGAAGATG.............................................................................................................................................................................................................. | 28 | 1 | 4.00 | 4.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGATGCACTGCTTGACCAGAATACTCAC................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGAAGATGAAGACACCATACCTGGATAT....................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................AACATCCTGAGGAAGATGGTGCAGGT...................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGATGCACTGCTTGACCAGAATACTC..................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGATGAAGATGAAGACACCATACCTGG........................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGATGAAGATGAAGACACCATACCTGGA.......................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGCACTGCTTGACCAGAATACTCACCCC................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............ACAAACGACTTAACATCCTGAGGAAGATG.............................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ATGAAGACACCATACCTGGATATCTTGt.................................................................................................................................. | 28 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GAAGATGAAGACACCATACCTG............................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGATGAAGATGAAGACACCATACCTG............................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................ACCGTACTGGATGGGGTTGATGCACTG..................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TACCTGGATATCTTGCAAACCGTACTG....................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................TCCCTGTCTCCCGCTGGGTGGTCTGATG................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................CGACTTAACATCCTGAGGAAGATGGTGCAG........................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................CGACTTAACATCCTGAGGAAGATGGTG........................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................ATGAAGATGAAGACACCATACCTGGATA........................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................AACGACTTAACATCCTGAGGAAGATGGTG........................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................TGGGTGGTCTGATGAAGATGAAGACA..................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................CCTTTTCCCTGTCTCCCGCTGGGTGG........................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................AACCGTACTGGATGGGGTTGATGCAC....................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................TTGACCAGAATACTCACCCCAGGaa........................................................................... | 25 | aa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AGATGAAGACACCATACCTGGATATCT..................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ATGCACTGCTTGACCAGAATACTCAC................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CCCTGTCTCCCGCTGGGTGGTCTGAT.................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................ACTGGATGGGGTTGATG.......................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATGAAGACACCATACCTGGATATC...................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GAAGACACCATACCTGGATATCT..................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGATGCACTGCTTGACCAGAATACTCA.................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TCCTGAGGAAGATGGTGCAGGTCTGt.................................................................................................................................................................................................. | 26 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................GATGAAGATGAAGACACCA.................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGATGCACTGCTTGACCAGAATAC....................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................GTGGTCTGATGAAGATGA.......................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGCAAACCGTACTGGATGGGGTTGA............................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TGGATGGGGTTGATGCACTGCTTG................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................TGCACTGCTTGACCAGAATACTCACCCCA............................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TCTGGCCTTTTCCCTGTCTCCCGC............................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TCTTGCAAACCGTACTGGATGGGattg............................................................................................................. | 27 | attg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................TCCCTGTCTCCCGCTGGGTGGTCTGATa................................................................................................................................................................. | 28 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................AGACACCATACCTGGATA........................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TACTGGATGGGGTTGATGCACTGCT................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TAGGGTAGAGAAAGAATAGAGAATGG........... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................TGAGGAAGATGGTGCAGGTCTGGCCT............................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................AACGACTTAACATCCTGAGGAAGA................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAGATGCCATCACTCACAAACGACTTAACATCCTGAGGAAGATGGTGCAGGTCTGGCCTTTTCCCTGTCTCCCGCTGGGTGGTCTGATGAAGATGAAGACACCATACCTGGATATCTTGCAAACCGTACTGGATGGGGTTGATGCACTGCTTGACCAGAATACTCACCCCAGGTGAGGGAGCCCCAGGGACTCTGAAGGGAGGCCTGGTAGTCTAGGGTAGAGAAAGAATAGAGAATGGAAGGAAGGTTA ..................................................................................(((.....)))..(((...(((....)))......((((((((.((.....)).))))..))))...))).................................................................................................. ..................................................................................83.......................................................................156............................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|