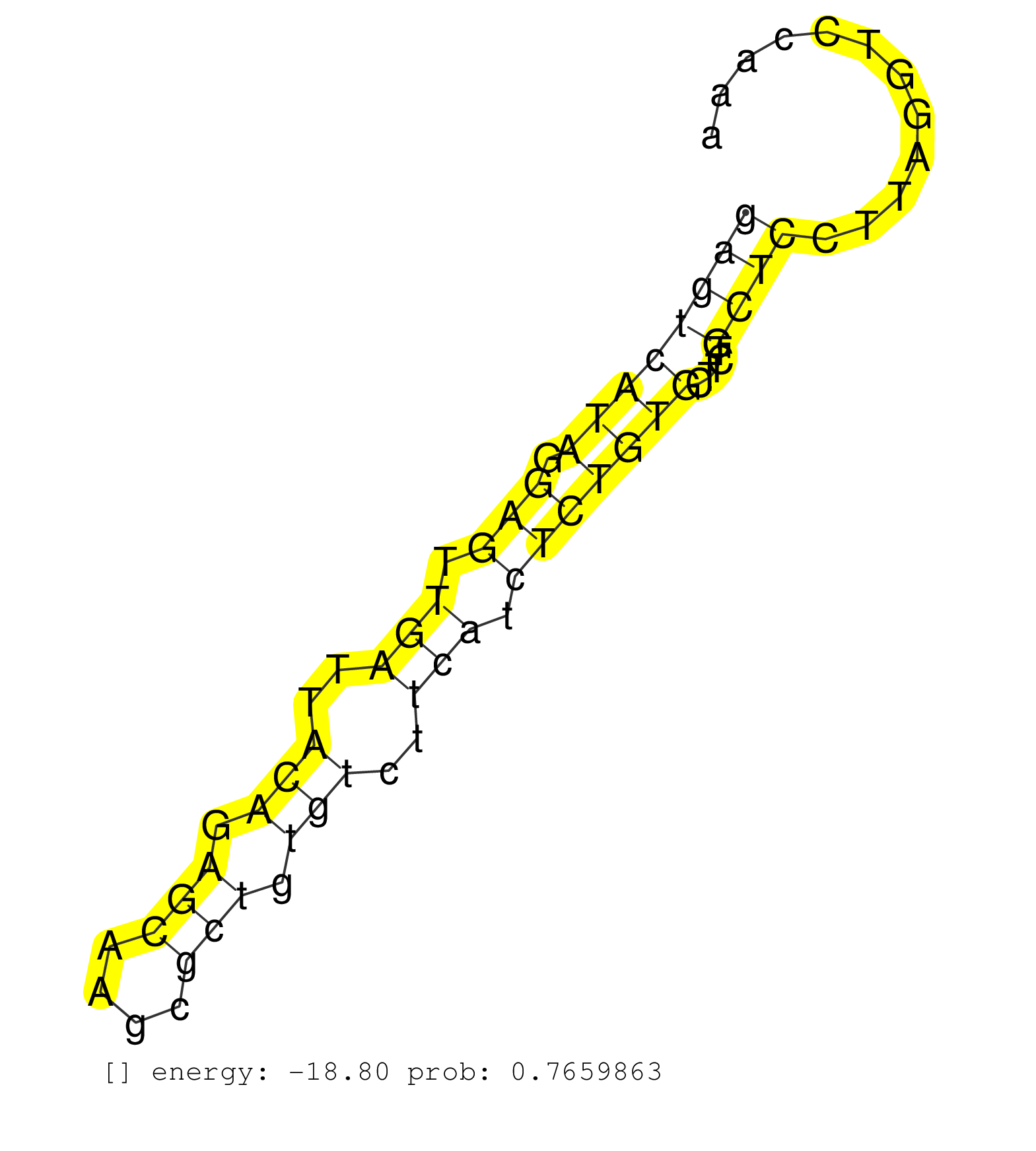
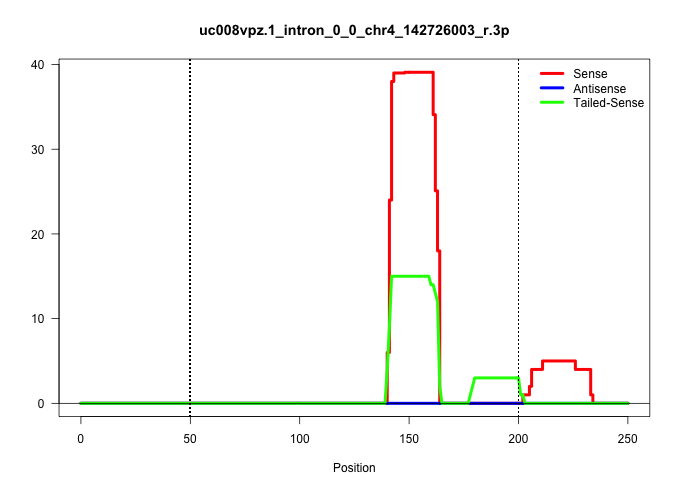
| Gene: Prdm2 | ID: uc008vpz.1_intron_0_0_chr4_142726003_r.3p | SPECIES: mm9 |
 |
 |
|
(8) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(1) PIWI.mut |
(26) TESTES |
| TGATGTGTCTATAGAGTGTTGCCTGGTCCTTAGCAGTCTTCTACATTTGTTAATTAAGTTAATGGAAGGGATTTGCACCCAGCTCAACCTCCAAATGAAATAATTTTGTTCACATATCTTAGCAGCTTCTAGCAATCGAGTCATAGGAGTTGATTACAGAGCAAGCGCTGTGTCTTCATCTCTGTGCTTCTGCTCCTTAGGTCCAAAAGAAGAGGATGAGAGGCCTTTGGCTTCTGCACCTGAGCAGCCA .........................................................................................................................................((((((((.(((.(((..(((.(((....))).)))..))).))))))).....))))....................................................... .........................................................................................................................................138..................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................ATAGGAGTTGATTACAGAGCAA...................................................................................... | 22 | 1 | 13.00 | 13.00 | - | 3.00 | 2.00 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................CATAGGAGTTGATTACAGAGC........................................................................................ | 21 | 1 | 11.00 | 11.00 | 2.00 | - | - | - | - | - | - | - | 3.00 | 1.00 | - | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................ATAGGAGTTGATTACAGAGCA....................................................................................... | 21 | 1 | 9.00 | 9.00 | - | 1.00 | - | - | 2.00 | - | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................CATAGGAGTTGATTACAGAGCAA...................................................................................... | 23 | 1 | 7.00 | 7.00 | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................ATAGGAGTTGATTACAGAGCAt...................................................................................... | 22 | t | 5.00 | 9.00 | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CATAGGAGTTGATTACAGAGCAt...................................................................................... | 23 | t | 5.00 | 3.00 | 1.00 | - | - | - | - | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TCATAGGAGTTGATTACAGAGC........................................................................................ | 22 | 1 | 5.00 | 5.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................CATAGGAGTTGATTACAGAGCA....................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................TCATAGGAGTTGATTACAGAG......................................................................................... | 21 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................ATAGGAGTTGATTACAGAGCAAa..................................................................................... | 23 | a | 2.00 | 13.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................AAGAAGAGGATGAGAGGCCTTTGGCTT................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CATAGGAGTTGATTACAGAG......................................................................................... | 20 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTCTGTGCTTCTGCTCCTTAGa................................................. | 23 | a | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................................................................................AAAGAAGAGGATGAGAGGCCTTTGGCTT................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTGTGCTTCTGCTCCTTAGaaa............................................... | 23 | aaa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TCATAGGAGTTGATTACAGAGCAtt..................................................................................... | 25 | tt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTCTGTGCTTCTGCTCCTTAGt................................................. | 22 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TCATAGGAGTTGATTACAGAGCAt...................................................................................... | 24 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................CCAAAAGAAGAGGATGAGAGGCCT........................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TCATAGGAGTTGATTACAGAGg........................................................................................ | 22 | g | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TAGGAGTTGATTACAGAGCAA...................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................GAGGATGAGAGGCCTTTGGCTTC................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................ATAGGAGTTGATTACAGAGCt....................................................................................... | 21 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTCTGTGCTTCTGCTCCTTAGa................................................. | 22 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TCATAGGAGTTGATTAtaga.......................................................................................... | 20 | taga | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GTTGATTACAGAGCA....................................................................................... | 15 | 12 | 0.08 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGATGTGTCTATAGAGTGTTGCCTGGTCCTTAGCAGTCTTCTACATTTGTTAATTAAGTTAATGGAAGGGATTTGCACCCAGCTCAACCTCCAAATGAAATAATTTTGTTCACATATCTTAGCAGCTTCTAGCAATCGAGTCATAGGAGTTGATTACAGAGCAAGCGCTGTGTCTTCATCTCTGTGCTTCTGCTCCTTAGGTCCAAAAGAAGAGGATGAGAGGCCTTTGGCTTCTGCACCTGAGCAGCCA .........................................................................................................................................((((((((.(((.(((..(((.(((....))).)))..))).))))))).....))))....................................................... .........................................................................................................................................138..................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|