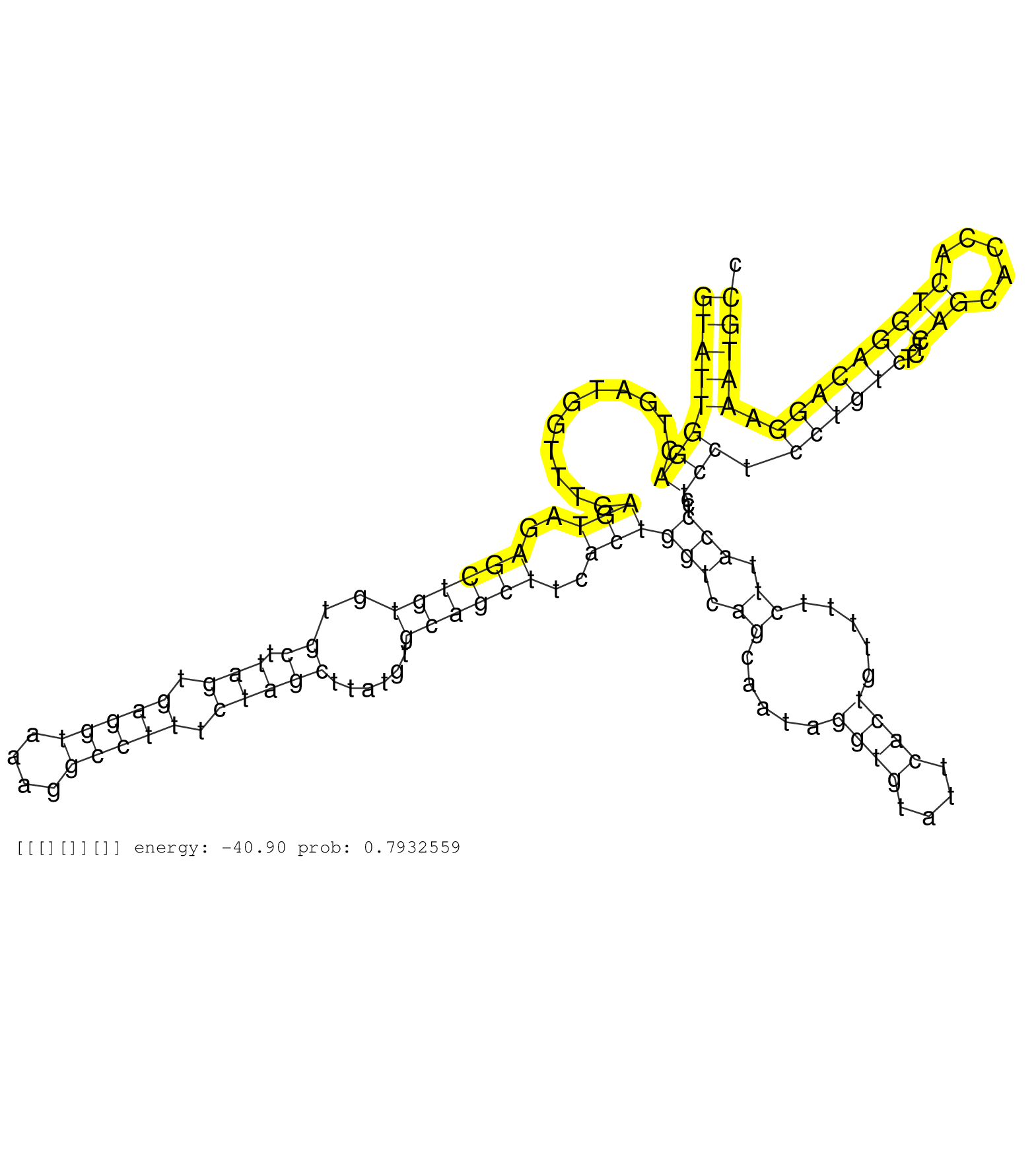

| Gene: Ddi2 | ID: uc008vpd.1_intron_1_0_chr4_141245247_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(24) TESTES |
| TGCAGACCAAGAATTAGCAGAAGCCATTCAAAAATCAGCCGAGGATGCAGGTATTGGACTGATGGTTTGAGTAGAGCTGTGTGCTTAGTGAGGTAAAGGCCTTTCTAGCTTATGTGCAGCTTCACTGGTCAGCAATAGGTGTATTCACTGTTTTCTTACCTCCTCCTCCTGTCTCTCAGCACCACTGGACAGGAAATGCCACTCGATAATAGAGTGCTTAGCACTGGGCTTAGTCCCCAGTACCACAGAC ..................................................((((((((...........(((..((((((..((.(((.(((((....))))).)))))......))))))..)))(((.((.....((((....)))).....)).)))...))).((((((...(((.....))))))))).)))))................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATTGGACTGATGGTTTGAGTAGAGC............................................................................................................................................................................. | 27 | 1 | 72.00 | 72.00 | 1.00 | 1.00 | 22.00 | - | 14.00 | - | 11.00 | - | 5.00 | 3.00 | 5.00 | 4.00 | 1.00 | - | 2.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - |
| ..................................................GTATTGGACTGATGGTTTGAGTAGAG.............................................................................................................................................................................. | 26 | 1 | 14.00 | 14.00 | 1.00 | - | 2.00 | - | 3.00 | - | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TTCAAAAATCAGCCGAGGATGCAGagc..................................................................................................................................................................................................... | 27 | agc | 13.00 | 0.00 | 3.00 | 4.00 | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................GTAGAGCTGTGTGCTTAGTGAG.............................................................................................................................................................. | 22 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATTGGACTGATGGTTTGAGTAGA............................................................................................................................................................................... | 25 | 1 | 4.00 | 4.00 | - | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATTGGACTGATGGTTTGAGTAGAGCT............................................................................................................................................................................ | 28 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................GCCGAGGATGCAGGTATTGGACTGA............................................................................................................................................................................................ | 25 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TATTGGACTGATGGTTTGAGTAGAGC............................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................TTCAAAAATCAGCCGAGGATGCAGag...................................................................................................................................................................................................... | 26 | ag | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .GCAGACCAAGAATTAGCAGAAGCCATT.............................................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TTCAAAAATCAGCCGAGGATGCAGaga..................................................................................................................................................................................................... | 27 | aga | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TCTCAGCACCACTGGACAGGAAATGC................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................AGCCGAGGATGCAGGTATTGGACTGA............................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .GCAGACCAAGAATTAGCAGAAGCCA................................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...AGACCAAGAATTAGCAGAAG................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................GGATGCAGGTATTGGACTGATGGTT....................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TTAGCAGAAGCCATTCAAAAATCAGCCGA................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TAGCAGAAGCCATTCAAAAATCAGCCGAGt.............................................................................................................................................................................................................. | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TTCAAAAATCAGCCGAGGATGCAGagaa.................................................................................................................................................................................................... | 28 | agaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................TCAAAAATCAGCCGAGGATGCAGagc..................................................................................................................................................................................................... | 26 | agc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TGTGCAGCTTCACTGGTCAGCAATAGGT.............................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................GAGTAGAGCTGTGTGCTTAGTGcggc............................................................................................................................................................ | 26 | cggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..CAGACCAAGAATTAGCAGAAGCCATTCAAA.......................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................ATTCAAAAATCAGCCGAGGA............................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................GAGGATGCAGGTATTGGACTGATGGTT....................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TAGCAGAAGCCATTCAAAAATCAGCC.................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTATTGGACTGATGGTTTGAGTA................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................TCAAAAATCAGCCGAGGATGCAGaga..................................................................................................................................................................................................... | 26 | aga | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TATTGGACTGATGGTTTGAGTAGAGCT............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GCAGAAGCCATTCAAAAATCAGCCGAG............................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................GATGCAGGTATTGGACTGATGGTTTGAGT.................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............TAGCAGAAGCCATTCAAAAATCAGCCGt................................................................................................................................................................................................................ | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................CATTCAAAAATCAGCCGAGGATGCAGagc..................................................................................................................................................................................................... | 29 | agc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................ATTGGACTGATGGTTTGAGTAGAGCT............................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGTAGAGCTGTGTGCTTAGTGAGGTAt.......................................................................................................................................................... | 27 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATTGGACTGATGGTTTGAGTAGAGCTG........................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| TGCAGACCAAGAATTAGCAGAAGCCATTCAAAAATCAGCCGAGGATGCAGGTATTGGACTGATGGTTTGAGTAGAGCTGTGTGCTTAGTGAGGTAAAGGCCTTTCTAGCTTATGTGCAGCTTCACTGGTCAGCAATAGGTGTATTCACTGTTTTCTTACCTCCTCCTCCTGTCTCTCAGCACCACTGGACAGGAAATGCCACTCGATAATAGAGTGCTTAGCACTGGGCTTAGTCCCCAGTACCACAGAC ..................................................((((((((...........(((..((((((..((.(((.(((((....))))).)))))......))))))..)))(((.((.....((((....)))).....)).)))...))).((((((...(((.....))))))))).)))))................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|