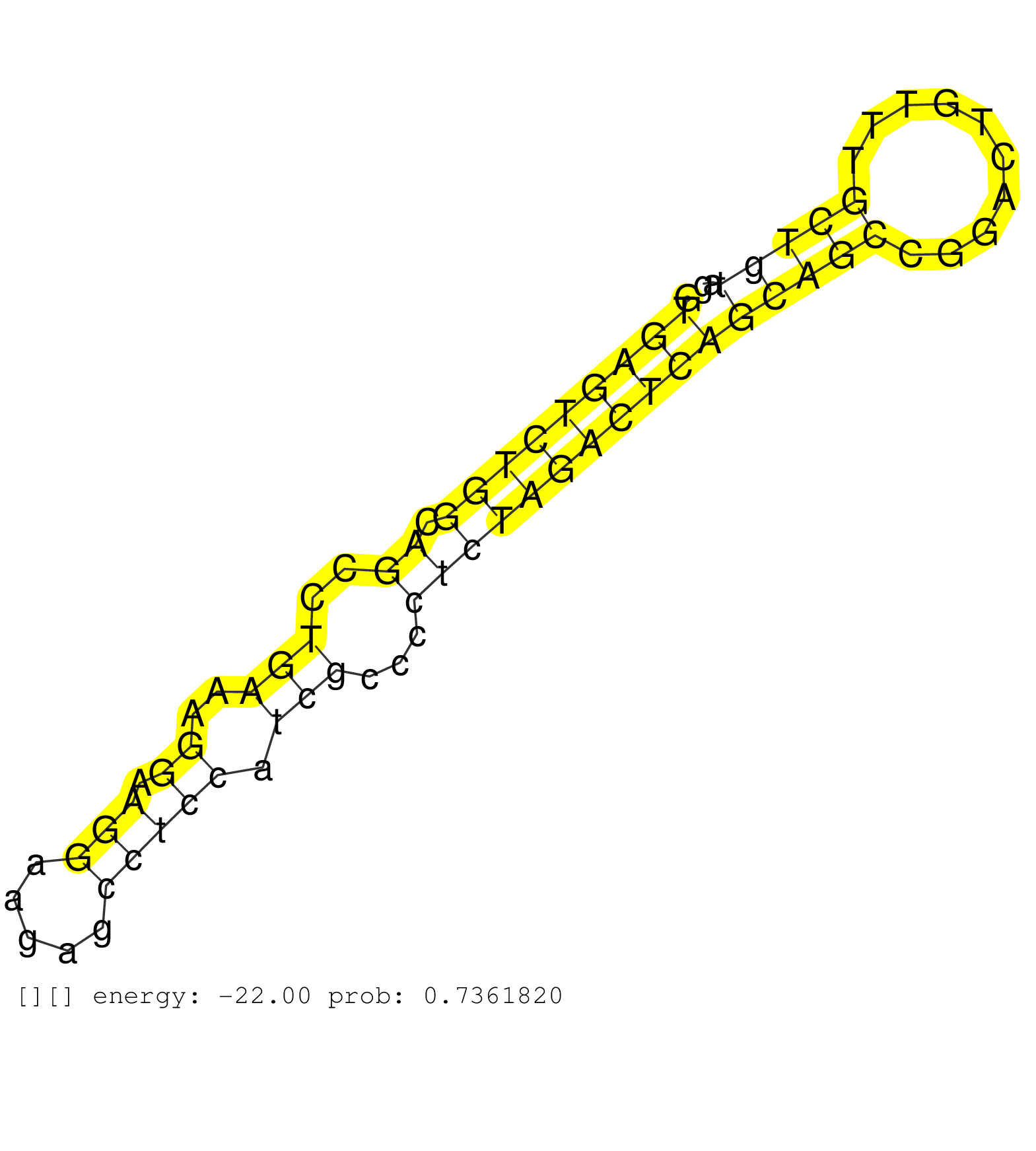

| Gene: Zubr1 | ID: uc008vmk.1_intron_29_0_chr4_138996457_f | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(26) TESTES |
| AGGAAAGCCAGGAACAGAGTGAGGTGGACCATGGAGATTTTGAGATGGTGGTGAGTCTGGCAGCCTGAAAGGAAGGAAGAGCCTCCATCGCCCCTCTAGACTCAGCAGCCGGACTGTTTGCTGTAGGACACTGATGGGGCTTTCTCTCTGCAGTCTGAGTCGATGGTCCTGGAAACAGCTGAAAATGTCAACAATGGCAACCC ...................................................(((((((((.((..(((..((.(((.....))))).)))...)))))))))))(((((..........)))))............................................................................... ..................................................51.........................................................................126........................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTCTGGCAGCCTGAAAGGAAGG............................................................................................................................... | 26 | 1 | 26.00 | 26.00 | 5.00 | 1.00 | 4.00 | 1.00 | 1.00 | 1.00 | 2.00 | - | 1.00 | 2.00 | 1.00 | - | 1.00 | 1.00 | 2.00 | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - |
| ..................................................GTGAGTCTGGCAGCCTGAAAGGAAGGA.............................................................................................................................. | 27 | 1 | 14.00 | 14.00 | 3.00 | - | 3.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTCTGGCAGCCTGAAAGGAAG................................................................................................................................ | 25 | 1 | 13.00 | 13.00 | 2.00 | 2.00 | - | 3.00 | - | - | - | 2.00 | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTCTGGCAGCCTGAAAGGAAGGAA............................................................................................................................. | 28 | 1 | 4.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGTCTGGCAGCCTGAAAGGAA................................................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGAGTGAGGTGGACCATGGAGATTT................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................TGAAAGGAAGGAAGAGCCTCCATCGCCCC............................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TCTGAGTCGATGGTCCTGGAAACAGCTG...................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................TAGACTCAGCAGCCGGACTGTTTGCT................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGTAGGACACTGATGGGGCTTTC........................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TAGACTCAGCAGCCGGACTGTTTGCTG................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GAGTGAGGTGGACCATGGA........................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTCTGGCAGCCTGAAAGGtagg............................................................................................................................... | 26 | tagg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GAGTGAGGTGGACCATGGAGA...................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........AGGAACAGAGTGAGGTGGACCATGGAGATTTT.................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGAGTGAGGTGGACCATGGAGATT.................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................CGATGGTCCTGGAAACAGC........................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................TGAAAGGAAGGAAGAGCCTCCATCGCC............................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................GAGATTTTGAGATGGTG......................................................................................................................................................... | 17 | 2 | 1.00 | 1.00 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTCTGGCAGCCTGAAAGGAAGGAAt............................................................................................................................ | 29 | t | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................GTGGACCATGGAGATTTTGAG............................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................TAGACTCAGCAGCCGGACTGTTTGC.................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TCCATCGCCCCTCTAGACTCAGCAGCCGG........................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTCTGGCAGCCTGAAAGGAAGGAA............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CGATGGTCCTGGAAACAGCTGAAAAc................. | 26 | c | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................GAGTCGATGGTCCTGGAAACA.......................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TGGAAACAGCTGAAAATGTC.............. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GAGTGAGGTGGACCATGG......................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............ACAGAGTGAGGTGGACCATGG......................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTCTGGCAGCCTGAAAGGAAGGt.............................................................................................................................. | 27 | t | 1.00 | 26.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGAGTCGATGGTCCTGGAAACAGC........................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................GATGGTCCTGGAAACAGCTGAAAATGTCAA............ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTGAGTCTGGCAGCCTGA....................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................GTGGACCATGGAGATTTTGA................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TAGACTCAGCAGCCGGACTGTTTGCTGT............................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............CAGAGTGAGGTGGACCATGGAG....................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............CAGAGTGAGGTGGACCA............................................................................................................................................................................ | 17 | 3 | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGAAAGCCAGGAACAGAGTGAGGTGGACCATGGAGATTTTGAGATGGTGGTGAGTCTGGCAGCCTGAAAGGAAGGAAGAGCCTCCATCGCCCCTCTAGACTCAGCAGCCGGACTGTTTGCTGTAGGACACTGATGGGGCTTTCTCTCTGCAGTCTGAGTCGATGGTCCTGGAAACAGCTGAAAATGTCAACAATGGCAACCC ...................................................(((((((((.((..(((..((.(((.....))))).)))...)))))))))))(((((..........)))))............................................................................... ..................................................51.........................................................................126........................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|