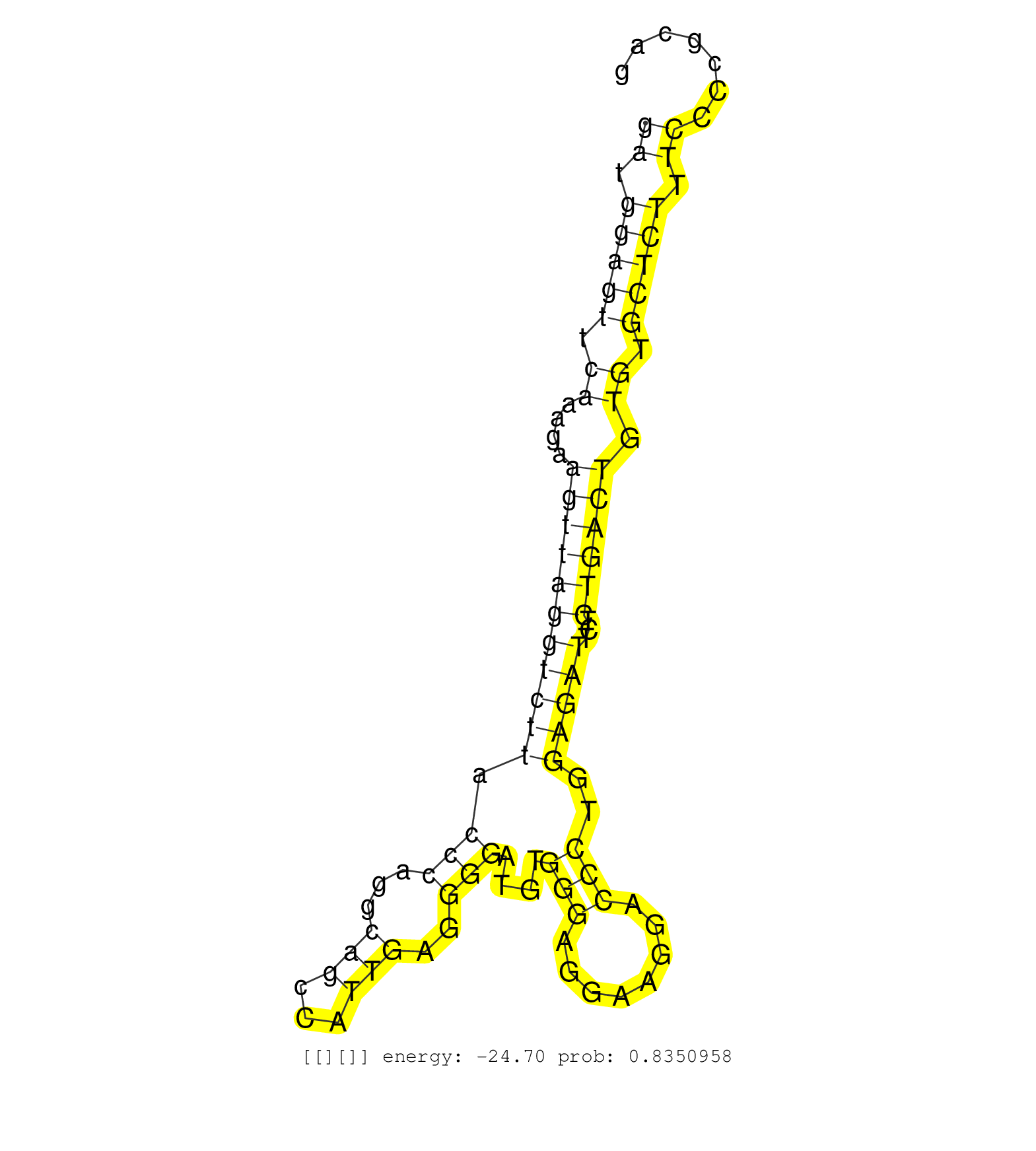

| Gene: Zubr1 | ID: uc008vmk.1_intron_19_0_chr4_138985571_f | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(3) PIWI.mut |
(21) TESTES |
| AGATAAAGCACAGGCTGAACTCCACTGGCATGTACGTGGCTAACACCAAGGTGAGCAGCACACGGGGCTGGCTCACTTTGTGGGTGGCACTCTTGTGAGCAGCGTTGTTGAACAAGTTGTGCCCATCTCCTCAGTATGAGGCTTGAATCCAGAGCCTTGCACATGCAAGGCAAGGACTCCCATCATTGAGGGGTCGGGCTGGCTTCACAGCTTCCTTTAGAATCAGACATAGAATATGTATTTGAAATTGCTCACTTCTGGTCAAATGATCCTTGAATAAATCAACCTTCTTTCATATGCTGAAGGTCTTGAGTCCCCCAGGCTGTTAGAAGGGTTATTGAGTACAGGGCACCATTTGATAGTTTGAGCTCCGGGGAGGAGCCTAGCATTCTCCCTCTCATATCTATGACACGATTAGGATGGAGTTCAAAGAAGTTAGGTCTTACCCAGGCAGCCATTGAGGGGATGTGGGAGGAAGGACCCTGGAGATTCTCTGACTGTGTGCTCTTTCCCCGCAGCCTGGCGGCTTCACCATCGAGATCAGTAACAACAGCAGCACCATGGTGAT ..................................................................................................................................................................................................................................................................................................................................................................................................................................((.(((((.((....(((((((((((.(((...(((...)))..)))....(((........)))..)))))...)))))).)).))))).))......................................................... ..................................................................................................................................................................................................................................................................................................................................................................................................................................419................................................................................................518................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................CATTGAGGGGATGTGGGAGGAAGGACCCTGGAGATTCTCTGACTGTGTGCTC............................................................. | 52 | 1 | 25.00 | 25.00 | 25.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCGAGATCAGTAACAACAGCAGCACCATG..... | 29 | 1 | 7.00 | 7.00 | - | - | 1.00 | 2.00 | - | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGATTCTCTGACTGTGTaga.............................................................. | 20 | aga | 5.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGATTCTCTGACTGTGTagag............................................................. | 21 | agag | 4.00 | 0.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................................CATTGAGGGGTCGGGCTGGCTTCA......................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCGAGATCAGTAACAACAGCAGCACC........ | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGGCATGTACGTGGCTAACACCAAGGctg.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 29 | ctg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....AAAGCACAGGCTGAACTCCACTGGCAT......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TAACAACAGCAGCACCATGGTG.. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TAAAGCACAGGCTGAACTCCACTGGCAT......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCTCTGACTGTGTGactg............................................................ | 18 | actg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................ACTCTTGTGAGCActtg............................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 17 | cttg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........CACAGGCTGAACTCCACTGGCA.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................CACTTTGTGGGTGGCACTCTTGTGAG..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........CAGGCTGAACTCCACTGGCAT......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........CACAGGCTGAACTCCACTGGCATGTAC..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCGAGATCAGTAACAACAGCAGCACCA....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........ACAGGCTGAACTCCACTGGCATGTACGT................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................CCACTGGCATGTACGT................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....AAAGCACAGGCTGAACTCCACTGGCAa......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........AGGCTGAACTCCACTGGCATGTACGT................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................CACTGGCATGTACGTGGCTAACACC......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........AGGCTGAACTCCACTGGCATGTACGTGGC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| AGATAAAGCACAGGCTGAACTCCACTGGCATGTACGTGGCTAACACCAAGGTGAGCAGCACACGGGGCTGGCTCACTTTGTGGGTGGCACTCTTGTGAGCAGCGTTGTTGAACAAGTTGTGCCCATCTCCTCAGTATGAGGCTTGAATCCAGAGCCTTGCACATGCAAGGCAAGGACTCCCATCATTGAGGGGTCGGGCTGGCTTCACAGCTTCCTTTAGAATCAGACATAGAATATGTATTTGAAATTGCTCACTTCTGGTCAAATGATCCTTGAATAAATCAACCTTCTTTCATATGCTGAAGGTCTTGAGTCCCCCAGGCTGTTAGAAGGGTTATTGAGTACAGGGCACCATTTGATAGTTTGAGCTCCGGGGAGGAGCCTAGCATTCTCCCTCTCATATCTATGACACGATTAGGATGGAGTTCAAAGAAGTTAGGTCTTACCCAGGCAGCCATTGAGGGGATGTGGGAGGAAGGACCCTGGAGATTCTCTGACTGTGTGCTCTTTCCCCGCAGCCTGGCGGCTTCACCATCGAGATCAGTAACAACAGCAGCACCATGGTGAT ..................................................................................................................................................................................................................................................................................................................................................................................................................................((.(((((.((....(((((((((((.(((...(((...)))..)))....(((........)))..)))))...)))))).)).))))).))......................................................... ..................................................................................................................................................................................................................................................................................................................................................................................................................................419................................................................................................518................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................................................................................................................CAGGCTGTTAGAAGttga............................................................................................................................................................................................................................................ | 18 | ttga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |